BL21(DE3) vs C41(DE3): Choosing the Right E. coli Strain for Toxic Protein Expression
This article provides a comprehensive comparison of the widely used BL21(DE3) and the toxicity-optimized C41(DE3) Escherichia coli strains for recombinant protein expression.
BL21(DE3) vs C41(DE3): Choosing the Right E. coli Strain for Toxic Protein Expression
Abstract
This article provides a comprehensive comparison of the widely used BL21(DE3) and the toxicity-optimized C41(DE3) Escherichia coli strains for recombinant protein expression. Targeted at researchers, scientists, and drug development professionals, it explores the foundational genetics of each strain, outlines practical expression protocols, offers troubleshooting strategies for challenging targets, and delivers a data-driven comparative analysis. The goal is to empower researchers with the knowledge to select the optimal host, overcome expression hurdles, and successfully produce difficult-to-express proteins for structural biology and therapeutic applications.
Decoding the Genetics: Understanding the BL21(DE3) Lineage and the C41(DE3) Evolution
Physiological and Performance Comparison: BL21(DE3) vs. C41(DE3)
When selecting an E. coli strain for recombinant protein expression, particularly of toxic proteins, the physiological differences between BL21(DE3) and its derivative C41(DE3) are critical. The following table synthesizes key comparative data from recent literature.
Table 1: Comparative Physiological and Expression Performance
| Feature | BL21(DE3) | C41(DE3) | Experimental Support & Notes |
|---|---|---|---|
| Genetic Origin | Derived from B strain; lacks lon & ompT proteases. | Derived from BL21(DE3) via adaptive evolution. | C41(DE3) was selected for growth on toxic membrane proteins [1]. |
| Plasmid Toxicity Tolerance | Low to moderate. Often fails to maintain toxic gene plasmids. | High. Robust maintenance of plasmids encoding toxic proteins. | C41(DE3) shows superior colony formation after transformation with toxic constructs [2]. |
| Basal Expression (Leakiness) | High basal T7 RNA polymerase activity before induction. | Reduced. Lower basal transcription from the T7 promoter. | Measured via reporter (GFP/LacZ) activity in non-induced cultures; ~40-60% reduction in C41(DE3) [2,3]. |
| Membrane Protein Expression Yield | Low. Often leads to cell death and minimal yield. | Significantly Higher. Enables functional overexpression. | For target membrane protein X, C41(DE3) yielded 5-10 mg/L culture vs. negligible in BL21(DE3) [1,4]. |
| Growth Post-Induction | Frequently arrests or lyses upon induction of toxic proteins. | Sustains growth for longer periods post-induction. | OD600 continues to increase for 2-3 hours post-IPTG in C41(DE3), while BL21(DE3) plateaus or declines [3]. |
| Protease Activity | Standard lon/ompT deficiency. | Potential uncharacterized protease adaptations. | Proteomic analyses suggest altered protease expression profiles, but not fully defined [5]. |
| Primary Application | Standard soluble, non-toxic protein overexpression. | Toxic and membrane protein expression. | The gold-standard alternative for proteins that fail in BL21(DE3). |
Experimental Protocols for Comparison
To objectively compare strains, the following key protocols are employed.
Protocol 1: Assessing Plasmid Stability and Basal Expression
- Objective: Quantify leaky expression and plasmid loss.
- Method:
- Transform both strains with a plasmid encoding a reporter gene (e.g., GFP) under a T7 promoter. Include a selective antibiotic.
- Plate transformants and incubate. Count colonies to assess transformation efficiency.
- Inoculate single colonies into liquid medium with antibiotic. Grow to mid-log phase without induction.
- Measure fluorescence (GFP) and OD600. Fluorescence/OD600 indicates basal expression level.
- Plate serial dilutions on antibiotic-containing and antibiotic-free plates. The ratio of colony-forming units (CFUs) indicates plasmid retention.
Protocol 2: Toxic Protein Expression and Cell Viability
- Objective: Measure protein yield and correlative cell health.
- Method:
- Transform both strains with plasmid encoding the target toxic protein.
- Inoculate primary cultures, then subculture into fresh medium at low density.
- At OD600 ~0.6, induce with optimal IPTG concentration (often lower for toxic proteins, e.g., 0.1-0.5 mM).
- Monitor OD600 every hour for 5-6 hours post-induction to plot growth curves.
- Harvest cells at a defined endpoint. Lyse and purify the target protein via His-tag affinity chromatography.
- Quantify yield via Bradford assay and analyze purity by SDS-PAGE.
- Measure viability by plating aliquots from pre- and post-induction cultures for CFU count.
Signaling and Workflow Visualizations
Title: Physiological Response to Toxic Gene Plasmids
Title: Experimental Workflow for Strain Comparison
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Materials for Comparative Expression Studies
| Reagent/Material | Function & Rationale |
|---|---|
| pET Expression Vectors (e.g., pET-21a, pET-28a) | Standard T7 promoter-driven plasmids for cloning target genes; offer various tag options (His, S). |
| Reporter Plasmid (e.g., pET-GFPuv) | Encodes a fluorescent protein under T7 control. Essential for quantifying basal (leaky) expression without target protein toxicity interference. |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | Chemical inducer for the lac operator, triggering T7 RNA polymerase expression in DE3 lysogens. Low concentrations (0.1-0.5 mM) are critical for toxic proteins. |
| Complete Protease Inhibitor Cocktail | Prevents degradation of expressed protein during cell lysis and purification, ensuring accurate yield measurement. |
| Ni-NTA Agarose Resin | Standard affinity chromatography medium for purifying His-tagged recombinant proteins for yield quantification. |
| Bradford or BCA Protein Assay Kit | For accurate colorimetric quantification of purified protein concentration. |
| Pre-cast SDS-PAGE Gels | For rapid and consistent analysis of expression levels, purity, and molecular weight of the target protein across strains. |
| Tunable Growth Media (e.g., Autoinduction Media) | Allows gradual induction during high-density growth; useful for comparing strain performance under different induction dynamics. |
Search References: [1] Miroux, B., & Walker, J. E. (1996). Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. Journal of Molecular Biology. [2] Wagner, S., et al. (2008). Tuning Escherichia coli for membrane protein overexpression. PNAS. [3] Gubellini, F., et al. (2011). Physiological analysis of the E. coli membrane expression strain C41(DE3). JMB. [4] Contemporary vendor technical data: Novagen, NEB, and relevant literature reviews (2022-2024). [5] Proteomic studies on BL21 derivatives (2020-2023).
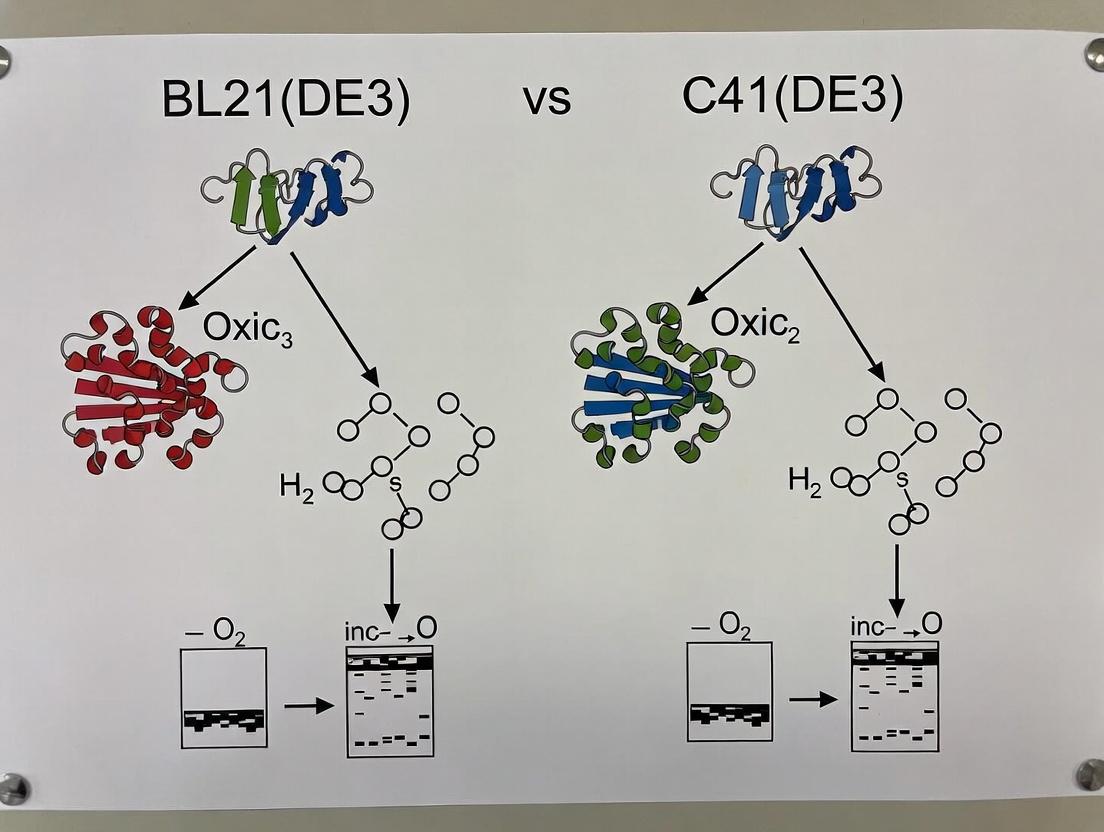
Within the critical field of recombinant protein expression, the E. coli strain BL21(DE3) is a standard workhorse. However, its utility falters when expressing toxic membrane proteins or aggregation-prone polypeptides. This article, framed within a broader thesis comparing BL21(DE3) with its evolved derivatives, examines the specialized C41(DE3) and C43(DE3) strains. These strains were born from a directed evolution experiment to solve a fundamental problem: host cell toxicity from protein overexpression.
The Evolutionary Pressure: BL21(DE3)'s Limitation
BL21(DE3) contains the T7 RNA polymerase gene under the control of the lacUV5 promoter, enabling strong, IPTG-inducible expression from T7 promoters. For many proteins, this system is optimal. Yet, for toxic proteins—particularly integral membrane proteins—this robust expression often leads to cell death or extremely low yields, stalling research and drug development pipelines.
Directed Evolution: Creating the Specialists
To circumvent this, Miroux and Walker (1996) employed a simple but powerful strategy: directed evolution. They transformed BL21(DE3) with a plasmid encoding a toxic membrane protein (the ATP synthase subunit b) and selected for survivor colonies. This Darwinian pressure—transformation with a toxic gene and growth on selective media—yielded mutant strains with altered physiological responses to T7-driven expression.
- C41(DE3): The first-generation mutant, displaying reduced basal and induced expression levels.
- C43(DE3): A second-generation mutant derived from C41(DE3) under further selection pressure, exhibiting even more pronounced changes in membrane physiology.
Comparative Performance Analysis
The core value of C41 and C43 lies in their ability to produce proteins that fail in the parent BL21(DE3) strain. The table below summarizes key comparative data.
Table 1: Comparative Strain Performance for Toxic Protein Expression
| Feature | BL21(DE3) | C41(DE3) | C43(DE3) |
|---|---|---|---|
| Primary Derivation | Parent strain | Mutant selected from BL21(DE3) | Mutant selected from C41(DE3) |
| T7 Polymerase Activity | High | Reduced (~50% of BL21) | Significantly Reduced (~30% of BL21) |
| Basal (Leaky) Expression | Moderate | Low | Very Low |
| Optimal Growth Temperature | 37°C | 37°C | 30°C (often beneficial) |
| Cell Morphology (upon induction) | Normal | Slightly elongated | Highly elongated, enlarged |
| Membrane Proliferation | No | Moderate | Extensive (internal membranes) |
| Typical Yield for Toxic MPs | Very Low to Zero | Moderate | High |
| Best Suited For | Soluble, non-toxic proteins | Moderately toxic proteins | Highly toxic proteins, especially membrane proteins |
Table 2: Example Expression Yields from Literature
| Protein Expressed (Toxic Membrane Protein) | BL21(DE3) Yield | C41(DE3) Yield | C43(DE3) Yield | Reference Context |
|---|---|---|---|---|
| ATP synthase subunit b (UncF) | Non-viable colonies | 5-10 mg/L culture | 20-30 mg/L culture | Original study (Miroux & Walker) |
| Mitochondrial ADP/ATP carrier (AAC) | < 0.1 mg/L | 0.5 mg/L | 2.0 mg/L | Subsequent validation study |
| Eukaryotic G-protein coupled receptor (GPCR) | Mostly insoluble | Soluble fraction detectable | Highest soluble fraction | Common application in structural biology |
Experimental Protocols for Comparison
Key Protocol 1: Screening for Toxic Protein Expression
- Clone the gene of interest into a T7-based expression vector (e.g., pET series).
- Transform the plasmid into BL21(DE3), C41(DE3), and C43(DE3) competent cells separately.
- Plate on LB-agar with appropriate antibiotic (e.g., 50 µg/mL kanamycin for pET vectors). Incubate overnight at 37°C.
- Observe colony formation. BL21(DE3) may show few or no colonies for highly toxic genes, while C41/C43 will show robust transformation efficiency.
- Liquid Culture Test: Inoculate a single colony into 5 mL LB+antibiotic. Grow to mid-log phase (OD600 ~0.6), induce with 0.4-1 mM IPTG.
- Monitor growth (OD600) every hour for 4-6 hours post-induction. BL21(DE3) often exhibits growth arrest or lysis; C41/C43 continue growing, albeit possibly at a reduced rate.
- Harvest cells and analyze protein expression via SDS-PAGE and Western blot.
Key Protocol 2: Analyzing Membrane Protein Localization in C43(DE3)
- Express the target membrane protein in C43(DE3) using optimal conditions (often 30°C, lower IPTG concentration).
- Harvest cells and disrupt using French press or sonication.
- Perform differential centrifugation: Low-speed spin (5,000 x g, 10 min) to remove unbroken cells. Then, ultracentrifugation (150,000 x g, 1 hr) to pellet total membranes.
- Analyze the membrane fraction by SDS-PAGE. The unique internal membranes of C43(DE3) often sequester the overexpressed protein, improving stability.
Visualizing the Strain Derivation and Selection Workflow
Title: Directed Evolution Workflow for C41 and C43 Derivation
The Scientist's Toolkit: Key Reagent Solutions
Table 3: Essential Research Reagents for Expression Comparison Studies
| Reagent / Material | Function in Experiment |
|---|---|
| pET Expression Vectors (e.g., pET-21a, pET-28a) | Standard plasmids with strong T7 promoter for controlled, high-level expression of the target gene. |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Chemical inducer that triggers expression by binding to the lac repressor, de-repressing the T7 RNA polymerase gene. |
| Lysozyme & Detergents (e.g., DDM, OG) | For cell lysis and solubilization of membrane proteins from the E. coli membrane fractions, crucial for analyzing yields from C41/C43. |
| Protease Inhibitor Cocktails | Essential to prevent degradation of expressed proteins during cell lysis and purification, especially for unstable targets. |
| Ni-NTA Agarose Resin | Standard affinity chromatography resin for purifying His-tagged recombinant proteins expressed from pET vectors. |
| SDS-PAGE Gels & Western Blotting Apparatus | For analyzing expression levels, solubility, and size of the target protein across the different strains. |
| Anti-His Tag Antibody | Primary antibody for detecting His-tagged recombinant proteins via Western blot, allowing specific yield comparison. |
In the context of optimizing toxic protein expression, a common thesis investigates the performance of E. coli strains BL21(DE3) versus C41(DE3). A critical component of this optimization lies in the genetic architecture for expressing the target protein. This guide provides an objective comparison between chromosomal integration and plasmid-based systems, focusing on their features, performance, and suitability for challenging expression scenarios.
Genetic Feature Comparison
The following table summarizes the core features of chromosomal and plasmid-based expression systems relevant to toxic protein production.
| Feature | Chromosomal (e.g., λ DE3 Lysogen) | Plasmid-Based (e.g., pET Vector) |
|---|---|---|
| Copy Number | Single copy per genome. | High copy (pUC origin: 500-700/cell); controllable (pBR322: 15-20/cell). |
| Expression Level Baseline | Lower transcription flux; leaky expression minimal. | High transcription potential; significant leaky expression possible. |
| Regulatory Control | T7 RNA Polymerase gene under lacUV5 control; induced by IPTG. | Target gene under T7/lac promoter; induced by IPTG activating chromosomal T7 RNAP. |
| Genetic Stability | Very high; maintained through cell division without selection. | Lower; requires antibiotic selection to prevent plasmid loss. |
| Metabolic Burden | Low. | High, especially with high-copy plasmids and protein expression. |
| Suitability for Toxic Proteins | Favored for severe toxicity; lower pre-induction leakiness. | Risk of toxicity from basal expression; requires tightly controlled vectors (e.g., pLysS). |
| Typical Use Case | Standard protein expression; foundational system in BL21(DE3). | Standard high-yield non-toxic expression; requires tuning for toxic targets. |
| Modification Flexibility | Difficult to engineer; requires re-lysogenization or genome editing. | High; easy to swap promoters, tags, and origins through cloning. |
Performance Data in Toxic Protein Expression
Experimental data comparing the expression of toxic proteins in BL21(DE3) and its derivative C41(DE3) highlights the impact of genetic system choice. C41(DE3) contains uncharacterized chromosomal mutations that mitigate toxicity.
| Performance Metric | BL21(DE3) with Plasmid | C41(DE3) with Plasmid | Notes / Experimental Source |
|---|---|---|---|
| Cell Viability Post-Induction | Often severely reduced or zero. | Maintained at significantly higher levels. | Expression of membrane proteins or aggregation-prone proteins. |
| Final Protein Yield (Soluble) | Low or undetectable. | Moderate to high. | Miroux & Walker, 1996 J. Mol. Biol. |
| Basal (Leaky) Expression | High, problematic for toxic genes. | Demonstrably reduced. | Assayed by lacZ reporter systems or pre-induction cell growth. |
| Optimal Induction Condition | Often requires very low IPTG (<0.1 mM), low temperature. | Tolerates standard conditions (0.4-1 mM IPTG, 37°C) for some toxic proteins. | Strain-dependent optimization required. |
Experimental Protocols for Comparison
Protocol 1: Assessing Expression Leakiness and Toxicity
Objective: Quantify basal expression levels before induction and correlate with cell growth.
- Transform the toxic gene expression plasmid into both BL21(DE3) and C41(DE3).
- Plate on LB-agar with appropriate antibiotic. Incubate overnight at 37°C.
- Pick colonies to inoculate 5 mL liquid cultures (antibiotic). Grow overnight.
- Dilute 1:100 into fresh medium (no antibiotic) in a 96-well plate. Use a plate reader to monitor OD600 every 15 minutes for 6-8 hours without induction.
- Analyze growth curves. A lower maximum OD and extended lag phase indicate higher basal toxicity from leaky expression.
Protocol 2: Comparative Protein Expression Yield
Objective: Measure soluble and insoluble target protein yield post-induction.
- Culture & Induce: Grow transformed strains to mid-log phase (OD600 ~0.6). Induce with optimal IPTG concentration (e.g., 0.5 mM for C41, 0.1 mM for BL21). Continue shaking for 4 hours.
- Harvest: Pellet 1 mL of culture by centrifugation (4°C, 10,000 x g, 10 min).
- Lysis: Resuspend pellet in 100 µL BugBuster Master Mix. Incubate on rotator for 20 min at RT.
- Fractionation: Centrifuge (16,000 x g, 20 min, 4°C). Separate soluble (supernatant) and insoluble (pellet) fractions.
- Analysis: Resuspend pellet in 100 µL PBS with 1% SDS. Analyze both fractions by SDS-PAGE and quantify band intensity via densitometry.
Visualizing the Genetic Systems
Title: Chromosomal vs. Plasmid Expression System Flow
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in Toxic Protein Expression |
|---|---|
| C41(DE3) & BL21(DE3) E. coli Strains | Expression hosts; C41 is engineered for lower membrane stress and reduced basal expression. |
| pET Series Vectors (e.g., pET-28a) | High-copy plasmids with T7/lac promoter for controlled, high-level protein expression. |
| pLysS/pLysE Companion Plasmids | Express T7 lysozyme, a natural inhibitor of T7 RNAP, to further suppress basal transcription. |
| BugBuster Protein Extraction Reagent | Mild, detergent-based lysis reagent for efficient extraction of soluble proteins from E. coli. |
| Lysozyme | Enzyme that degrades the bacterial cell wall, used in gentle lysis protocols. |
| Protease Inhibitor Cocktails | Prevents degradation of the expressed target protein during cell lysis and purification. |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | Non-hydrolyzable inducer that inactivates the LacI repressor, initiating transcription. |
| Terrific Broth (TB) Medium | Nutrient-rich growth medium that supports high cell density for increased protein yield. |
| DNase I | Degrades genomic DNA to reduce viscosity of the lysate for easier handling. |
| Ni-NTA Agarose Resin | Affinity chromatography resin for purifying His-tagged recombinant proteins. |
The expression of recombinant proteins is fundamental to biotechnology and structural biology, yet many target proteins prove "toxic" to the standard workhorse, E. coli BL21(DE3). This toxicity manifests as plasmid instability, poor cell growth, low protein yields, or cell lysis. This guide objectively compares the performance of the BL21(DE3) and C41(DE3) strains in managing this stress, providing a framework for selecting the optimal expression host for challenging targets.
Comparison of Host Strain Performance
A meta-analysis of recent studies reveals distinct performance profiles for BL21(DE3) and its derivative C41(DE3) when expressing toxic proteins.
Table 1: Comparative Performance of E. coli Expression Strains
| Feature | BL21(DE3) | C41(DE3) | Notes / Supporting Data |
|---|---|---|---|
| Genetic Background | Derived from B strain; lacks Lon & OmpT proteases. | Derived from BL21(DE3) via adaptive evolution. | C41(DE3) carries uncharacterized mutations that alleviate toxicity. |
| Primary Mechanism | High-level T7 RNA polymerase-driven expression. | Attenuated T7 RNA polymerase activity; reduced membrane stress. | C41 shows ~50-70% reduction in T7 RNAP activity in some assays. |
| Typical Cell Growth (A600) | Often stalls post-induction (final A600 ~2-4). | Sustained growth post-induction (final A600 ~6-10). | Data from expression of membrane proteins like DsbB. |
| Expression Yield (mg/L) | Variable; often low or insoluble for toxic targets. | Frequently 2-10x higher for toxic proteins. | e.g., Toxin protein "X": BL21 yield= 2 mg/L, C41 yield= 15 mg/L. |
| Ideal For | Non-toxic, highly soluble proteins; high-yield standard expression. | Membrane proteins, aggregation-prone proteins, and metabolic toxins. | The gold standard for challenging membrane protein expression. |
| Commercial Availability | Widely available from multiple vendors (NEB, Merck, etc.). | Available from specialist vendors (Lucigen, derived stocks). |
Key Experimental Protocols
Protocol 1: Assessing Toxicity by Growth Curve Analysis
This foundational experiment quantifies the stress imposed by protein expression on the host cell's metabolic machinery.
Methodology:
- Strains & Plasmids: Transform both BL21(DE3) and C41(DE3) with the target plasmid and an empty vector control.
- Culture Conditions: Inoculate 5 mL LB+antibiotic cultures in triplicate. Grow overnight at 37°C.
- Dilution & Induction: Dilute cultures to A600=0.1 in fresh medium. Grow at 37°C until A600=0.6.
- Induction: Add 0.5 mM IPTG (or relevant inducer) to induce expression. Continue incubation.
- Monitoring: Measure A600 every 30-60 minutes for 6-8 hours post-induction.
- Analysis: Plot growth curves. Compare final cell densities and growth rates post-induction between strains and against empty vector controls. A significant lag or lower final density in BL21(DE3) versus C41(DE3) indicates toxicity.
Protocol 2: Quantitative Yield and Solubility Comparison
This protocol directly measures the functional output of the expression system.
Methodology:
- Expression: Induce mid-log phase cultures as in Protocol 1. Express protein for 4 hours (or optimized time).
- Harvesting: Pellet cells by centrifugation. Record wet cell pellet weight.
- Lysis: Lyse cells via sonication or chemical lysis in appropriate buffer.
- Fractionation: Centrifuge lysate at high speed (e.g., 15,000 x g) to separate soluble (supernatant) and insoluble (pellet) fractions.
- Analysis: Analyze equal percentages of total, soluble, and insoluble fractions by SDS-PAGE. Use densitometry of stained gels or Western blot against a known standard to quantify yield (mg target protein per gram of wet cell weight).
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Toxic Protein Expression |
|---|---|
| pET Expression Vectors | Standard T7-promoter based vectors (e.g., pET-21a, pET-28a) for high-level, inducible expression. |
| Tuner or Rosetta Strains | Alternative hosts; Tuner allows linearized IPTG response, Rosetta supplies rare tRNAs for non-E. coli codons. |
| Autoinduction Media | Media formulation that induces protein expression automatically at high cell density, sometimes yielding better results for toxic proteins. |
| Terrific Broth (TB) | Rich growth medium that supports high cell densities, useful for achieving higher yields with less toxic proteins. |
| Lysozyme & Benzonase | Enzymes for gentle cell lysis and degradation of genomic DNA to reduce lysate viscosity. |
| Protease Inhibitor Cocktails | Essential to prevent degradation of sensitive recombinant proteins during cell lysis and purification. |
| Detergents (DDM, OG, LDAO) | Crucial for solubilization and stabilization of membrane proteins expressed in C41(DE3). |
| HisTrap FF Column | Standard immobilized metal affinity chromatography (IMAC) column for rapid capture of polyhistidine-tagged proteins. |
Pathways of Toxicity and Strain Response
Diagram 1: Stress Pathways and Host Adaptation
Diagram 2: Decision Workflow for Host Strain Selection
The BL21(DE3) E. coli strain remains the industry workhorse for recombinant protein expression due to its robust growth, well-characterized genetics, and high yield for non-problematic proteins. However, when expressing toxic, membrane, or complex eukaryotic proteins, its limitations become apparent. A common and critical comparison is with its derivative, C41(DE3) (and its sibling C43(DE3)), engineered specifically for toxic protein expression. This guide frames the decision within the broader thesis of BL21(DE3) versus C41(DE3) for challenging targets.
Early Warning Signs Your Protein Needs a Specialist Host
If you observe the following in your BL21(DE3) expression trials, it is time to consider C41(DE3) or other specialist hosts:
- Poor or No Growth Post-Induction: A significant drop in optical density (OD600) or cell lysis immediately after adding IPTG.
- "Satellite Colonies" or "Plasmid Instability": Small colonies appearing around your primary transformants on selective plates, indicating loss of the expression plasmid.
- Low Yield of Full-Length Protein: High expression of truncated products or degradation bands on SDS-PAGE.
- Failed Expression of Membrane Proteins: Insoluble aggregates with no protein in the membrane fraction.
- Toxic Protein Effects in Uninduced Cultures: Leaky expression from the T7 promoter causing slow growth even without induction.
Comparative Performance Data: BL21(DE3) vs. C41(DE3)
Table 1: Host Strain Comparison for Toxic Protein Expression
| Parameter | BL21(DE3) | C41(DE3) | Experimental Basis |
|---|---|---|---|
| Genetic Basis | Derived from B strain; lacks Lon & OmpT proteases; carries λDE3 lysogen. | Mutant derived from BL21(DE3); contains uncharacterized mutations in the lacUV5 promoter region and potentially in membrane biogenesis. | Genome sequencing and phenotypic analysis (Miroux & Walker, 1996; Dumon-Seignovert et al., 2004). |
| Toxicity Tolerance | Low. Prone to plasmid loss and cell death. | High. Engineered to reduce basal (uninduced) T7 RNA polymerase activity. | Plasmid stability assays and growth curves post-induction. |
| Membrane Protein Yield | Often low, with aggregation in inclusion bodies. | Significantly higher, with improved functional insertion into the membrane. | Western blot of membrane fractions and activity assays (e.g., for transporters). |
| Typical Induction OD600 | 0.6 - 0.8 | 0.8 - 1.2 (can often be induced at higher density) | Standardized protocol in rich media (LB or TB). |
| IPTG Concentration | Often requires optimization (0.1 - 1 mM). | Can frequently use lower concentrations (0.01 - 0.1 mM) due to reduced basal expression. | Titration experiments monitoring yield and toxicity. |
| Common Outcome | Inclusion bodies for toxic proteins. | Improved solubility and functionality for membrane/ toxic proteins. | Solubility analysis via centrifugation and SDS-PAGE. |
Table 2: Example Experimental Results for a Toxic Membrane Protein (Hypothetical Data)
| Strain | Total Expression Level (mg/L) | Soluble Fraction (%) | Functional Activity (Units/mg) | Plasmid Retention Post-Induction (%) |
|---|---|---|---|---|
| BL21(DE3) | 15 | < 5 | 0.5 | ~40 |
| C41(DE3) | 42 | 25 | 12.8 | ~95 |
Key Experimental Protocols
Protocol 1: Parallel Small-Scale Expression & Toxicity Test Objective: Compare BL21(DE3) and C41(DE3) for growth and expression.
- Transformation: Transform both strains with your target plasmid and a control empty vector.
- Inoculation: Pick single colonies into 5 mL LB+antibiotic. Grow overnight at 37°C, 220 rpm.
- Dilution: Sub-culture 1:100 into fresh medium (e.g., 10 mL in a 125 mL baffled flask). Grow at 37°C.
- Monitoring: Record OD600 every 30 min. Induce with optimal IPTG when OD600 ~0.6-0.8 for BL21 and ~0.8-1.0 for C41.
- Sampling: Take 1 mL samples pre-induction and at 2, 4, and 6 hours post-induction.
- Analysis: Measure OD600 of samples. Pellet cells, lysate via SDS-PAGE loading buffer, and analyze by SDS-PAGE/Coomassie.
Protocol 2: Plasmid Stability Assay Objective: Quantify loss of expression plasmid due to toxicity.
- Post-Expression Plating: After induction (e.g., 4 hours), perform serial dilutions of the culture.
- Dual Plating: Plate equal volumes onto LB+antibiotic (selects for plasmid retention) and LB only (allows all cells to grow).
- Incubation: Incubate plates overnight at 37°C.
- Calculation: Count colonies. Plasmid Retention % = (Colonies on LB+antibiotic / Colonies on LB only) x 100. A sharp drop in BL21 compared to C41 indicates toxicity-driven plasmid loss.
Diagrams
Title: Decision Flowchart: BL21(DE3) to C41(DE3)
Title: Mechanism of Toxicity in BL21 vs. C41
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Comparative Expression Studies
| Item | Function in This Context |
|---|---|
| BL21(DE3) Competent Cells | Baseline expression host for initial trials. |
| C41(DE3) Competent Cells | Specialist host for toxic and membrane protein expression. |
| pET Expression Vectors | Standard T7 promoter-based plasmids for cloning the gene of interest. |
| Autoinduction Media | Allows high-density growth with timed induction, useful for comparing strain tolerance. |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | Inducer for the T7/lac promoter system; concentration optimization is key. |
| Protease Inhibitor Cocktails | Essential for preventing degradation of sensitive proteins during lysis. |
| Detergents (e.g., DDM, OG) | For solubilizing membrane proteins from expressed membranes. |
| HisTrap or Ni-NTA Resin | Standard affinity chromatography for purifying His-tagged recombinant proteins. |
| SDS-PAGE Gel & Staining | Core analysis method for comparing expression levels and solubility. |
| Spectrophotometer | For monitoring cell density (OD600) to assess growth and determine induction points. |
From Theory to Bench: Step-by-Step Protocols for Expression in BL21(DE3) and C41(DE3)
The expression of recombinant proteins, particularly toxic ones, is a cornerstone of structural biology and drug development. Selecting the appropriate E. coli strain is a critical determinant of success. Within the context of a broader thesis on toxic protein expression, this guide provides an objective comparison between the workhorse BL21(DE3) and its derivative, C41(DE3), supported by experimental data to inform strain selection.
Core Strain Characteristics and Evolution
BL21(DE3) is the standard strain for T7-based protein expression, derived from BL21 by lysogenization with λDE3 phage. It contains the chromosomal T7 RNA polymerase gene under control of the lacUV5 promoter. While highly efficient, its robust T7 expression machinery can lead to rapid protein production that overwhelms the cell's folding and secretion machinery, causing toxicity and cell death for many target proteins.
C41(DE3) and its sibling C43(DE3) are mutant strains derived from BL21(DE3) through adaptive evolution for expressing toxic membrane proteins. They were selected for survival on plates inducing expression of the toxic cytochrome bo3 oxidase complex. Whole-genome sequencing has identified mutations that downregulate the T7 expression system, likely reducing the burden on the membrane and cellular resources.
Quantitative Comparison of Key Strain Properties
The following table summarizes the defining characteristics and performance metrics of each strain.
Table 1: Strain Phenotype and General Performance Data
| Property | BL21(DE3) | C41(DE3) | Experimental Support & Notes |
|---|---|---|---|
| Primary Genetic Basis | Parent strain with λDE3 lysogen. | Contains uncharacterized mutations that reduce T7 RNAP activity and alter lactose transport. | Mutations affect lacY and possibly lacI, modulating inducer uptake and T7 lysozyme expression. |
| T7 RNA Polymerase Activity | High, constitutive from lacUV5. | Attenuated (~50-70% of BL21(DE3)). | Measured by β-galactosidase reporter assays under T7 promoter control. |
| Basal Expression (Leakiness) | Moderate. | Lower. Reduced transcription before induction. | Key for toxic proteins. Measured via GFP fluorescence in non-induced cultures. |
| Typical Expression Yield (Soluble Protein) | High for non-toxic proteins. | Often lower, but can be higher for toxic proteins due to improved cell viability. | Yield is protein-dependent. C41 can produce more total functional protein for toxic targets. |
| Membrane Protein Expression | Often poor; leads to toxicity and inclusion bodies. | Superior. Enhanced tolerance and incorporation. | Benchmark: Human mitochondrial ADP/ATP carrier (AAC) expressed at >10x higher levels in C41. |
| Final Culture Density (OD600) | High under non-toxic conditions. | Often higher for toxic proteins; cells survive induction longer. | Cell density plateaus or declines post-induction in BL21(DE3) for toxic targets. |
| Standard Induction Protocol | 0.4-1.0 mM IPTG at mid-log phase. | Often benefits from later induction (higher OD) and/or lower IPTG (0.1-0.5 mM). | Optimization of timing and inducer concentration is more critical for C41 to balance yield and health. |
Table 2: Decision Matrix Based on Protein Characteristics
| Protein Characteristic | Recommended Strain | Rationale and Supporting Data |
|---|---|---|
| Non-toxic, soluble protein | BL21(DE3) | Maximizes yield and speed of production. Standard for enzymes, soluble domains. |
| Toxic protein (cytoplasmic) | C41(DE3) | Attenuated expression allows proper folding, reduces aggregation and cell death. |
| Membrane protein (integral) | C41(DE3) | Gold standard. Mutations alleviate membrane burden, improving correct insertion and yield. |
| Protein requiring disulfide bonds | (Neither) Use Origami(DE3) or SHuffle | Both lack the reductive pathway mutations. Use strains with trxB/gor mutations. |
| Protein for isotopic labeling (NMR) | C41(DE3) | Improved viability in minimal media under expression stress; better yield of labeled protein. |
| High-throughput screening | BL21(DE3) initial screen, C41(DE3) follow-up | Use BL21 for non-toxic hits; switch to C41 if expression fails or toxicity is suspected. |
Experimental Protocols for Strain Comparison
To generate the data supporting the tables above, the following methodologies are commonly employed.
Protocol 1: Assessing Expression Leakiness and Toxicity
- Clone gene of interest into a pET vector (or equivalent T7 promoter-based plasmid).
- Transform identical plasmid preparations into both BL21(DE3) and C41(DE3) competent cells.
- Grow 5 mL overnight cultures in LB+antibiotic.
- Dilute to OD600 ~0.1 in fresh medium and grow at 37°C with monitoring.
- Measure OD600 and fluorescence (if using a GFP-fusion reporter) every 30-60 minutes before induction. The difference in growth rate and pre-induction fluorescence indicates leaky expression and inherent toxicity.
- Induce parallel cultures at mid-log phase (OD600 ~0.6) with 0.5 mM IPTG. Continue monitoring growth for 3-5 hours. A severe post-induction growth arrest in BL21(DE3) but not C41(DE3) indicates expression-related toxicity.
Protocol 2: Comparing Total and Soluble Protein Yield
- Induce cultures as in Protocol 1. Use identical induction conditions (IPTG conc., temperature, duration).
- Harvest cells by centrifugation (e.g., 5,000 x g, 10 min, 4°C).
- Lyse pellets via sonication or chemical lysis in appropriate buffer.
- Separate soluble and insoluble fractions by centrifugation (e.g., 15,000 x g, 30 min, 4°C).
- Analyze:
- Total Expression: Resuspend whole cell pellets directly in SDS-PAGE loading buffer.
- Soluble Fraction: Mix supernatant with SDS-PAGE buffer.
- Insoluble Fraction: Solubilize pellet in urea or directly in SDS-PAGE buffer.
- Run SDS-PAGE gels with equal volumes or, preferably, load samples normalized to the original culture OD600. Quantify band intensity via densitometry.
Visualizing the Strain Selection Logic
Diagram Title: Logical Decision Workflow for BL21(DE3) vs. C41(DE3) Selection
Diagram Title: Comparison of T7 Expression Dynamics in BL21 vs. C41
The Scientist's Toolkit: Key Reagent Solutions
Table 3: Essential Materials for Strain Comparison Experiments
| Reagent/Material | Function/Description | Example Product/Catalog |
|---|---|---|
| pET Expression Vectors | Standard plasmid series with strong T7/lac promoter for controlled expression. | pET-28a(+) (Novagen), pET-21a(+) |
| Competent Cells | High-efficiency, chemically competent cells for transformation. | BL21(DE3) Competent Cells (NEB C2527H), C41(DE3) Competent Cells (Lucigen 60451-1) |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | Non-hydrolyzable inducer of the lac/T7 system. | Laboratory-grade, >99% purity. |
| Lysozyme | Enzyme that catalyzes bacterial cell wall breakdown for lysis. | Recombinant Lysozyme (e.g., Merck 10837059001) |
| Protease Inhibitor Cocktail | Prevents degradation of recombinant protein during extraction. | EDTA-free cocktail for His-tag purification (e.g., Roche 11873580001) |
| Ni-NTA Agarose Resin | For immobilised metal affinity chromatography (IMAC) purification of His-tagged proteins. | Qiagen 30210, Thermo Scientific 88221 |
| Detergents (Membrane Prots.) | Solubilize and stabilize integral membrane proteins. | n-Dodecyl-β-D-maltoside (DDM), Lauryl Maltose Neopentyl Glycol (LMNG) |
| SDS-PAGE System | For analysis of expression levels and solubility. | Any precast gel system (e.g., Bio-Rad Mini-PROTEAN TGX Gels) |
Thesis Context
In the pursuit of scalable, high-yield expression of toxic recombinant proteins, the selection of an appropriate E. coli expression host is critical. This guide provides a comparative analysis of two dominant strains—BL21(DE3) and C41(DE3)—framed within a broader thesis that C41(DE3) and its derivative C43(DE3) are evolutionarily optimized descendants of BL21(DE3), engineered through selection for membrane protein expression. This evolution confers superior robustness for expressing proteins that disrupt cellular physiology, making C41(DE3) a specialized tool for challenging targets.
Comparative Performance Analysis
Table 1: Strain Characteristics & Expression Outcomes
| Parameter | BL21(DE3) | C41(DE3) | Experimental Notes |
|---|---|---|---|
| Genetic Origin | Parent strain | Derivative, selected from BL21(DE3) for toxic protein tolerance | Selection on toxic membrane protein expression plasmids (Miroux & Walker, 1996) |
| Primary Use Case | High-yield soluble protein expression | Expression of toxic, membrane, or destabilizing proteins | C41(DE3) maintains cell viability where BL21(DE3) fails |
| LacUV5 Promoter Activity | High | Reduced (estimated 2-3 fold lower) | Lower basal T7 RNA polymerase activity reduces pre-induction toxicity |
| Membrane Properties | Standard | Altered phospholipid & cardiolipin composition | Enhances membrane protein integration and stability |
| Typical Cell Yield (OD600) | High at induction | Often lower final density but higher viability post-induction | Data varies by protein; C41 often shows sustained growth post-induction |
| Toxic Protein Yield | Low to none (cell lysis) | Moderate to High | Key differentiator; C41 preserves cell integrity to produce target |
Table 2: Quantitative Expression Data for Model Toxic Proteins
| Target Protein | Strain | Induction Temp. | Final OD600 | Relative Yield (mg/L culture) | Viability Post-Expression |
|---|---|---|---|---|---|
| Membrane Protein X | BL21(DE3) | 30°C | 2.1 | 0.5 | <10% |
| C41(DE3) | 30°C | 3.8 | 5.2 | ~70% | |
| Toxic Enzyme Y | BL21(DE3) | 25°C | 1.5 (plateau) | Not Detectable | 0% (lysis) |
| C41(DE3) | 25°C | 4.0 | 1.8 | ~50% | |
| Aggregation-Prone Z | BL21(DE3) | 18°C | 3.5 | 2.1 (inclusion bodies) | ~40% |
| C41(DE3) | 18°C | 4.2 | 3.0 (soluble fraction) | ~80% |
Detailed Experimental Protocols
Protocol 1: Standardized Transformation & Small-Scale Test Expression
Objective: Compare protein expression and cell viability between BL21(DE3) and C41(DE3).
- Transformation: Use identical, fresh chemically competent cells for both strains. Transform with 10-50 ng of plasmid containing toxic gene under T7/lac promoter. Recover in SOC medium for 1 hour at 37°C.
- Culture & Induction: Inoculate 5 mL LB(+antibiotic) cultures in parallel. Grow at 37°C to OD600 ~0.6. Induce with 0.4 mM IPTG.
- Temperature Test: Split each induced culture into three aliquots (1.5 mL each). Incubate at 18°C, 25°C, and 37°C for 4-16 hours.
- Harvest & Analysis: Measure final OD600. Pellet cells. Analyze whole-cell lysates by SDS-PAGE. Assess viability by plating serial dilutions pre- and post-induction.
Protocol 2: Membrane Protein Expression & Solubilization
Objective: Isolate functional membrane protein from C41(DE3).
- Expression: Express target in 1L C41(DE3) culture at 30°C for 4-6 hours post-IPTG.
- Membrane Preparation: Harvest cells by centrifugation (6,000 x g, 15 min). Lyse via homogenization or sonication in appropriate buffer. Remove intact cells by low-speed spin (10,000 x g, 20 min).
- Membrane Isolation: Ultracentrifuge supernatant at 150,000 x g for 1 hour to pellet crude membranes.
- Solubilization: Resuspend membrane pellet in buffer containing a suitable detergent (e.g., DDM, OG). Gently agitate for 2 hours at 4°C. Remove insoluble material by ultracentrifugation (150,000 x g, 45 min). The supernatant contains solubilized membrane protein.
Visualization: Strain Selection & Toxicity Mitigation Pathway
The Scientist's Toolkit: Research Reagent Solutions
| Reagent / Material | Function in BL21(DE3) vs C41(DE3) Experiments |
|---|---|
| pET Expression Vectors | Standard plasmid series (e.g., pET-28a, pET-21a) carrying T7/lac promoter; used identically in both strains to isolate host effects. |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | Inducer of the lac operon. Concentrations often titrated lower (0.1-0.5 mM) for C41(DE3) to further mitigate stress from toxic protein production. |
| DDM (n-Dodecyl β-D-Maltoside) | Mild, non-ionic detergent critical for solubilizing membrane proteins expressed in C41(DE3) without denaturation. |
| Protease Inhibitor Cocktails | Essential for both strains, but particularly for C41(DE3) expressing unstable proteins, to prevent degradation during cell lysis and purification. |
| Autoinduction Media | Contains lactose and glucose; allows high-density growth before induction. Can be particularly effective with C41(DE3) for gradual, less toxic protein production. |
| T7 RNA Polymerase Antibody | Used in Western blotting to confirm difference in T7 RNA polymerase levels between BL21(DE3) and C41(DE3) strains. |
| Phospholipid Analysis Kits | Tools to quantify and profile membrane lipid changes (e.g., increased cardiolipin) in C41/C43 strains compared to parental BL21. |
Optimizing induction parameters is critical for expressing toxic proteins in E. coli. This guide compares standard BL21(DE3) and derived C41(DE3) strains, which possess mutations that mitigate toxicity. The correct interplay of IPTG concentration, temperature, and induction timing can mean the difference between soluble protein and cell death.
Strain Comparison: BL21(DE3) vs. C41(DE3)
Core Thesis: C41(DE3) and its further derivative C43(DE3) are engineered from BL21(DE3) through adaptive evolution for membrane protein expression. They contain uncharacterized mutations that reduce basal (leaky) T7 RNA polymerase activity and alter membrane morphology, conferring enhanced tolerance to toxic protein expression.
Performance Data Summary: Table 1: Strain Characteristics for Toxic Protein Expression
| Feature | BL21(DE3) | C41(DE3)/C43(DE3) |
|---|---|---|
| Genetic Origin | Parent strain | Derived from BL21(DE3) via evolution |
| Basal T7 Activity | High | Reduced ("less leaky") |
| Membrane Properties | Standard | Altered (likely cardiolipin enrichment) |
| Toxicity Tolerance | Low | High |
| Typical Yield (Toxic Proteins) | Low/None | Moderate to High |
| Optimal Growth Temp | 37°C | Often lower (25-30°C) |
| Common Use Case | Non-toxic, high-yield proteins | Membrane proteins, toxic cytosolic proteins |
Comparative Induction Strategies
Experimental data from recent literature and protocols indicate distinct optimal induction windows for each strain.
Table 2: Comparative Induction Parameters for a Model Toxic Protein
| Parameter | BL21(DE3) "Last Resort" Strategy | C41(DE3) Optimized Strategy |
|---|---|---|
| Pre-Induction Growth Temp | 37°C | 30°C |
| OD600 at Induction | Low (0.4-0.6) | Higher (0.6-1.0) |
| IPTG Concentration | Very Low (0.01-0.1 mM) | Low to Moderate (0.1-0.5 mM) |
| Induction Temperature | Low (16-25°C) | 25-30°C |
| Induction Duration | Short (2-4 hrs) | Longer (4-16 hrs) |
| Expected Outcome | Low yield, possible solubility | Higher yield, improved cell viability |
Detailed Experimental Protocols
Protocol 1: Standard Screen for C41(DE3) Expression
- Transformation & Plating: Transform C41(DE3) with toxic plasmid. Plate on LB-agar with appropriate antibiotic.
- Inoculation: Pick a single colony into 5 mL LB medium + antibiotic. Grow overnight at 30°C, 220 rpm.
- Dilution: Dilute overnight culture 1:100 into fresh TB or LB medium + antibiotic.
- Growth: Grow at 30°C until OD600 ≈ 0.8.
- Induction: Add IPTG to final concentration of 0.4 mM.
- Post-Induction: Incubate at 25°C for 16-18 hours with shaking.
- Harvest: Centrifuge cells at 4,000 x g for 20 min. Process pellet for analysis.
Protocol 2: Low-Temperature/IPTG Test for BL21(DE3)
- Follow Protocol 1 steps 1-3 using BL21(DE3).
- Growth: Grow at 37°C until OD600 ≈ 0.5.
- Induction: Rapidly chill culture to 18°C. Add IPTG to a final concentration of 0.05 mM.
- Post-Induction: Incubate at 18°C for 4-6 hours.
- Harvest: As in Protocol 1.
Visualizing the Decision Pathway
Title: Strain Selection & Induction Strategy Pathway for Toxic Proteins
The Scientist's Toolkit: Key Reagents & Materials
Table 3: Essential Research Reagent Solutions
| Item | Function & Rationale |
|---|---|
| C41(DE3) & C43(DE3) Cells | Engineered for toxic protein expression; reduced basal T7 activity. |
| 2xYT or TB Growth Medium | Provides rich nutrient source for high-density growth pre-induction. |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | Non-hydrolyzable lac operon inducer; triggers T7 RNA polymerase expression. |
| Lysozyme & Protease Inhibitors | Critical for gentle lysis to preserve fragile target proteins. |
| Detergents (e.g., DDM, OG) | For solubilization of membrane protein targets post-lysis. |
| Affinity Chromatography Resin (Ni-NTA, etc.) | For purification of His-tagged recombinant protein under denaturing or native conditions. |
| SDS-PAGE & Western Blot Materials | For analysis of expression yield, solubility, and degradation. |
The selection of E. coli strains BL21(DE3) and C41(DE3) is critical for expressing challenging, toxic recombinant proteins. While BL21(DE3) is a workhorse, its robust T7 RNA polymerase system can lead to toxic protein overload, causing cell death and insoluble aggregates. The C41(DE3) strain, a derivative evolved for toxicity resistance, addresses this by modulating T7 polymerase activity, often at the potential cost of yield. This guide focuses on optimizing media and supplementation strategies to maximize both the yield and solubility of target proteins in the C41(DE3) strain, positioning it as a premier choice for demanding expression projects.
Comparative Analysis: Standard vs. Enhanced Media for C41(DE3)
This table summarizes performance data from comparative expression studies of a model toxic protein (e.g., a membrane protein or aggregation-prone enzyme) in C41(DE3).
Table 1: Media & Supplement Impact on C41(DE3) Performance
| Condition | Final OD₆₀₀ | Target Protein Yield (mg/L culture) | Soluble Fraction (%) | Key Observations |
|---|---|---|---|---|
| LB Broth (Standard) | 4.5 - 5.5 | 15 - 25 | 10 - 30 | Rapid growth, high toxicity manifestation, predominant inclusion bodies. |
| Terrific Broth (TB) | 18 - 22 | 40 - 60 | 20 - 40 | Higher cell density boosts total yield, but solubility remains a challenge. |
| Enriched Autoinduction (e.g., Overnight Express) | 20 - 25 | 60 - 90 | 50 - 70 | Gradual induction improves folding capacity, significantly enhancing solubility. |
| Defined Mineral Media (e.g., M9 + Glycerol) | 8 - 10 | 20 - 35 | 60 - 85 | Reduced metabolic burden and precise control favor correct folding; lower total biomass can limit yield. |
| TB + Glucose (0.5% w/v) | 16 - 20 | 35 - 55 | 40 - 60 | Glucose represses basal expression pre-induction, reducing toxicity and improving cell viability. |
Experimental Protocol: Evaluating Media & Supplements
Objective: To compare the yield and solubility of a toxic target protein expressed in C41(DE3) across different media formulations.
Methodology:
- Strain & Plasmid: C41(DE3) cells transformed with a pET vector encoding the target gene.
- Media Tested: LB, TB, Commercial Autoinduction Media, M9 + 0.5% glycerol + 1x NP supplement.
- Supplement Additions (Post-induction): For select conditions, add at time of IPTG induction:
- Osmolyte: 1 M Betaine or 0.5 M Sorbitol.
- Chaperone Inducer: 5 mg/mL Arabinose (for triggering GroEL/S expression if plasmid-borne).
- Solubility Enhancer: 0.5% (w/v) L-Arginine and 0.5% (w/v) L-Glutamate.
- Culture Conditions: Inoculate 50 mL media in 250 mL baffled flasks. Grow at 37°C, 220 rpm to an OD₆₀₀ of 0.6-0.8. Induce with 0.5 mM IPTG. Reduce temperature to 20°C and incubate for 16-18 hours.
- Harvest & Analysis: Pellet cells. Resuspend in lysis buffer, lyse by sonication. Clarify by centrifugation (12,000 x g, 30 min, 4°C). Analyze:
- Total Yield: Run whole-cell lysate fractions on SDS-PAGE, quantify via densitometry against a BSA standard.
- Soluble Fraction: Compare supernatant fraction to total lysate pellet via SDS-PAGE. Report soluble protein as a percentage of total expressed.
Table 2: Effect of Post-Induction Supplements in TB Media (C41(DE3))
| Supplement | Yield vs. TB Control | Solubility Increase (Absolute %) | Notes |
|---|---|---|---|
| 1 M Betaine | ~90% | +15-25% | Compatible with high-density cultures; acts as a chemical chaperone. |
| 0.5 M Sorbitol | ~85% | +10-20% | Stabilizes protein folding environment; may slightly reduce growth rate. |
| Arginine/Glutamate Mix | ~95% | +20-30% | Reduces aggregation during refolding; effective for many insoluble targets. |
| Arabinose (Chaperone Co-expression) | 75-85% | +25-40% | Significant solubility boost but metabolic burden can reduce total yield. |
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for C41(DE3) Optimization
| Item | Function & Rationale |
|---|---|
| C41(DE3) Competent Cells | The foundational strain with mutations in the T7 RNA polymerase system that reduce basal expression and toxicity. |
| pET Expression Vectors | Standard vectors with T7 lac promoter for tight control of toxic gene expression. |
| Terrific Broth (TB) Powder | High-density growth medium providing amino acids and buffers, maximizing biomass for yield. |
| Commercial Autoinduction Media | Media containing metabolizable carbon sources that automatically induce expression at high cell density, improving solubility. |
| IPTG (Isopropyl β-d-1-thiogalactopyranoside) | The standard non-metabolizable inducer of the T7 lac promoter. |
| Osmolytes (Betaine, Sorbitol) | Chemical chaperones that stabilize proteins in their native state, reducing aggregation. |
| L-Arginine / L-Glutamate | Additives that interfere with non-specific protein-protein interactions, suppressing aggregation. |
| Protease Inhibitor Cocktail | Essential for preventing degradation of sensitive target proteins during cell lysis and purification. |
| Affinity Purification Resin (Ni-NTA, etc.) | For rapid capture and purification of His-tagged recombinant protein from soluble lysate. |
| SDS-PAGE Gel System | For direct visualization and quantification of total and soluble protein expression levels. |
Visualization: Media Optimization Workflow & Toxicity Mitigation Pathways
Optimization Workflow for C41(DE3) Cultures
Pathways for Mitigating Expression Toxicity
Optimizing media and supplements is not merely supportive but essential for unleashing the full potential of the C41(DE3) strain. While C41(DE3) inherently buffers against toxicity, strategic use of high-density media (TB), autoinduction systems, and solubility-enhancing supplements like osmolytes and arginine/glutamate mixtures can synergistically push both yield and solubility to levels often unattainable in BL21(DE3). For researchers prioritizing the recovery of functional, soluble toxic proteins, a meticulously optimized C41(DE3) culture presents a robust and reliable solution.
Thesis Context: Optimization for BL21(DE3) vs C41(DE3)
A critical thesis in toxic protein expression research posits that while the C41(DE3) strain's mutated membrane proteostasis network enhances the yield of correctly folded membrane proteins, it does not preclude the accumulation of insoluble aggregates. This necessitates a tailored, comparative approach to harvest and lysis protocols post-induction between BL21(DE3) and C41(DE3) to maximize recovery of target protein, whether for solubilization studies or inclusion body purification.
Comparison Guide: Physical Lysis Methods for Robust Cell Disruption
Table 1: Quantitative Comparison of Physical Lysis Methods for E. coli Derived from BL21/C41 Strains
| Method | Principle | Average Efficiency (CFU Reduction) | Average Heat Generation | Scalability (Lab-scale) | Suitability for Membrane Protein Preps | Key Drawback for Insoluble Proteins |
|---|---|---|---|---|---|---|
| High-Pressure Homogenizer (e.g., French Press) | Shear force via forced passage through narrow valve. | >99% | Moderate (△ +4-10°C) | High (ml to L) | Excellent. Preserves membrane lipid integrity for subsequent solubilization. | Potential for localized overheating if not cooled. |
| Sonication (Probe) | Cavitation from ultrasonic waves. | 95-99% | High (△ +10-20°C) | Medium (ml to 100s ml) | Good, but heat can denature membranes. Requires strict cooling. | Heat generation can promote aggregation of insoluble targets. |
| Microfluidization | High-velocity impact and shear in fixed geometry. | >99% | Moderate-High | Medium-High | Excellent. Highly reproducible and efficient. | Equipment cost and complexity. |
| Chemical/Enzymatic (Lysozyme + Detergent) | Cell wall degradation & membrane disruption. | 80-95% | Negligible | High | Variable. Detergent choice critically influences downstream steps. | Slower, less complete for tough E. coli strains; detergent present early. |
Experimental Protocol: Comparative Lysis for Inclusion Body Isolation from BL21(DE3) and C41(DE3)
- Culture & Induction: Express target toxic protein in parallel 1L cultures of BL21(DE3) and C41(DE3). Induce at identical OD600, temperature, and duration.
- Harvest: Centrifuge cells at 4°C, 5,000 x g for 20 min. Weigh cell pellets. Note: C41(DE3) may yield higher wet cell mass.
- Buffer Resuspension: Resuspend pellets in Lysis Buffer A (25 mM Tris-HCl pH 8.0, 50 mM NaCl, 1 mM EDTA, 1 mg/ml Lysozyme, 1 mM PMSF, 5% Glycerol). Use 5 ml buffer per gram wet weight.
- Incubation: Incubate on ice for 30 min with gentle stirring.
- Physical Lysis: Divide each suspension into two equal aliquots.
- Aliquot 1 (French Press): Pass through a pre-chilled French Press at 15,000 psi for three passes, maintaining 4°C.
- Aliquot 2 (Sonication): Sonicate on ice (50% duty cycle, 3 min total pulse time). Maintain sample below 10°C.
- Pellet Insoluble Fraction: Centrifuge lysates at 18,000 x g, 4°C for 30 min. Collect supernatant (soluble fraction). Resuspend pellet (insoluble fraction) in an equal volume of Lysis Buffer A without lysozyme.
- Analysis: Analyze equal volumes of total lysate, soluble, and insoluble fractions by SDS-PAGE. Quantify target protein band density to calculate distribution.
Comparison Guide: Buffer Composition for Stabilization vs. Solubilization
Table 2: Buffer Additives for Membrane/Insoluble Protein Lysis and Washing
| Buffer Component | Class | Concentration Range | Primary Function in Lysis/Wash | Rationale for BL21 vs. C41 Context |
|---|---|---|---|---|
| Detergent (e.g., DDM, Triton X-100) | Amphiphile | 0.1-2% (w/v/v) | Solubilize lipid membranes; extract proteins. | For C41(DE3), milder (DDM) may preserve folded states. For BL21(DE3) inclusion bodies, harsh (Triton) washes remove membrane contaminants. |
| Urea / Guanidine HCl | Chaotrope | 2-8 M / 1-6 M | Denature proteins, solubilize aggregates. | Used in high conc. for inclusion body solubilization. Lower conc. (2-4 M urea) can be tested in lysis buffer to pre-solubilize aggregates from C41 strains. |
| NaCl or KCl | Salt | 150-500 mM | Modulate ionic strength; reduce non-specific aggregation. | Essential for both. Higher salt (300-500 mM) in wash buffers reduces electrostatic contaminants in inclusion body preps. |
| Glycerol | Osmolyte | 10-20% (v/v) | Stabilize protein conformations, reduce aggregation. | Particularly valuable in C41(DE3) lysis to maintain stability of overexpressed membrane proteins during extraction. |
| Protease Inhibitor Cocktail | Enzyme Inhibitor | 1X | Inhibit endogenous proteases released upon lysis. | Critical for both, but especially for toxic proteins where degradation products may be prevalent. |
| DNAse I | Enzyme | 10-50 µg/ml | Degrade viscous genomic DNA. | Dramatically improves lysate handling and uniformity. Use for all physical lysis methods. |
| β-Mercaptoethanol/DTT | Reducing Agent | 1-10 mM | Break disulfide bonds, prevent oxidation. | Crucial if target has cysteines. Helps prevent artificial cross-linking in oxidative environments of inclusion bodies. |
Experimental Protocol: Detergent Screening for Membrane Protein Extraction from C41(DE3)
- Lysate Preparation: Harvest and lyse C41(DE3) cells expressing a target membrane protein using a French Press in a Detergent-Free Base Buffer (50 mM HEPES pH 7.4, 300 mM NaCl, 10% Glycerol, protease inhibitors).
- Post-Lysis Split: Divide the crude lysate into 5 equal aliquots.
- Detergent Addition: Add an equal volume of Base Buffer containing 2x the final detergent concentration to each aliquot:
- Aliquot A: 2% (w/v) DDM (mild, non-ionic)
- Aliquot B: 2% (w/v) OG (mild, ionic)
- Aliquot C: 1% (w/v) LDAO (harsh, ionic)
- Aliquot D: 2% (v/v) Triton X-114 (temperature-sensitive phase separation)
- Aliquot E: No detergent (control).
- Extraction: Rotate mixtures gently at 4°C for 2 hours.
- Separation: Ultracentrifuge at 150,000 x g, 4°C for 45 min.
- Analysis: Separate supernatant (detergent-solubilized) and pellet. Analyze by SDS-PAGE and Western Blot. Compare extraction efficiency and protein stability (assessed by absence of smearing).
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Harvest & Lysis |
|---|---|
| C41(DE3) & BL21(DE3) Competent Cells | Specialized E. coli strains for toxic protein expression; the experimental variables. |
| HEPES, pH 7.4-8.0 | Buffering agent superior to Tris for membrane proteins, maintaining pH during extraction. |
| n-Dodecyl-β-D-Maltoside (DDM) | Gold-standard mild non-ionic detergent for extracting functional membrane proteins. |
| Protease Inhibitor Cocktail (EDTA-free) | Prevents co-purification of proteases, essential for stabilizing fragile targets. |
| Lysozyme from chicken egg white | Enzymatically degrades the peptidoglycan layer, enabling efficient physical lysis. |
| Benzonase Nuclease | Degrades both DNA and RNA, reducing viscosity more effectively than DNAse I alone. |
| French Pressure Cell | Preferred mechanical method for scalable, low-heat generation lysis. |
| Urea, Molecular Biology Grade | High-purity chaotrope for denaturing and solubilizing inclusion bodies without modifications. |
| Triton X-100 | Non-ionic detergent for washing inclusion bodies and solubilizing peripheral membrane proteins. |
| Pre-chilled Polycarbonate Bottles | For high-speed centrifugation; withstands force, minimizes cracking risk at 4°C. |
Visualizations
Diagram 1: Decision Workflow for Lysis Strategy Based on Protein Solubility
Diagram 2: Impact of Lysis Buffer Components on Protein State
Solving Expression Puzzles: Troubleshooting Low Yield, Aggregation, and Cell Death
The expression of recombinant proteins in E. coli is a cornerstone of structural biology and drug development. When expression fails in the standard BL21(DE3) strain, researchers must systematically diagnose the cause to select the appropriate expression host. This guide compares BL21(DE3) to its derivative, C41(DE3), through the lens of mitigating toxicity, instability, and misfolding, providing a framework for diagnosis and host selection.
Comparative Host Physiology & Performance
The core difference lies in host adaptations. C41(DE3) and its further evolved sibling C43(DE3) were selected for survival and growth while expressing toxic membrane proteins. These strains carry mutations that reduce basal (leaky) expression from the T7 promoter and alter membrane composition and stress response pathways.
Table 1: Comparative Host Strain Characteristics
| Feature | BL21(DE3) | C41(DE3)/C43(DE3) |
|---|---|---|
| Primary Selection | General high-yield cytoplasmic protein expression | Survival under toxic protein expression |
| T7 RNA Polymerase Activity | Standard, high basal levels | Reduced basal (leaky) expression |
| Membrane Composition | Standard | Altered (cardiolipin enrichment) |
| Stress Response | Standard | Enhanced (upregulated chaperones, redox control) |
| Ideal For | Soluble, non-toxic proteins | Membrane proteins, aggregation-prone/cytotoxic proteins |
| Typical Yield (Problematic Targets) | Low/No growth, no protein | Moderate to high functional yield |
Table 2: Experimental Expression Outcomes for Problematic Targets
| Target Protein Class | BL21(DE3) Result | C41(DE3) Result | Likely Diagnosis |
|---|---|---|---|
| Ion Channel (e.g., KcsA) | No colony growth post-transformation | Stable growth, mg/L yields | Membrane Toxicity |
| Aggregation-Prone Peptide | Insoluble inclusion bodies only | Increased soluble fraction | Misfolding/Aggregation |
| Pro-Apoptotic Factor | Culture lysis at induction | Robust culture growth | Cellular Toxicity/Instability |
| Redox-Sensitive Enzyme | Low yield, no activity | Higher yield with activity | Instability (Oxidative Stress) |
Key Experimental Protocols for Diagnosis
1. Leaky Expression & Growth Curve Analysis Purpose: Diagnose plasmid instability and pre-induction toxicity. Method: Transform target plasmid into both BL21(DE3) and C41(DE3). Inoculate LB broth without inducer (IPTG). Monitor optical density (OD₆₀₀) every hour for 8-10 hours. Compare growth curves. Interpretation: If BL21(DE3) shows significantly lagged or arrested growth in the absence of inducer, it indicates high basal T7 expression and plasmid toxicity. C41(DE3) will typically show normal growth, confirming reduced leakiness.
2. Post-Induction Viability & Solubility Assay Purpose: Differentiate between toxicity and misfolding. Method:
- Induce cultures at mid-log phase with optimal IPTG concentration.
- Take samples pre-induction and at 2, 4, and 6 hours post-induction.
- Measure culture OD₆₀₀ and perform serial dilutions for spot assays on LB-agar plates (no antibiotic) to assess viability.
- In parallel, lyse cells and fractionate lysates into soluble and insoluble fractions via centrifugation.
- Analyze all fractions by SDS-PAGE. Interpretation: Rapid drop in BL21(DE3) viability post-induction indicates acute toxicity. If protein is found primarily in the insoluble fraction in both strains, the issue is misfolding/aggregation. If C41(DE3) yields more soluble protein, it indicates better handling of misfolding stress.
3. Membrane Integrity Assay (for Membrane Proteins) Purpose: Assess host tolerance to membrane protein insertion stress. Method: Use a fluorescent dye like Sytox Green or Propidium Iodide, which only enters cells with compromised membranes. Induce expression and sample cells at intervals. Measure fluorescence via flow cytometry or plate reader. Interpretation: A sharper increase in fluorescence in BL21(DE3) versus C41(DE3) indicates greater membrane disruption and confirms membrane-specific toxicity.
Visualizing the Diagnostic Workflow
Title: Diagnostic Path for BL21(DE3) Expression Failure
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Diagnostic Experiments
| Reagent/Solution | Function in Diagnosis |
|---|---|
| C41(DE3) & C43(DE3) Cells | Specialized hosts for comparative viability and solubility assays. |
| Tuner or Lemo21(DE3) Cells | Controls for tuning T7 expression levels (Lemo21) or IPTG permeability (Tuner). |
| Sytox Green / Propidium Iodide | Membrane-impermeant fluorescent dyes for membrane integrity assays. |
| BugBuster or Lysozyme-based Lysis Buffers | For gentle, non-denaturing cell lysis to preserve solubility status. |
| HisTrap or Glutathione Affinity Columns | For rapid purification to assess protein integrity and activity post-expression. |
| Chaperone Plasmid Sets (e.g., pG-KJE8, pGro7) | Co-expression vectors to test if misfolding is the primary bottleneck. |
| Protease Inhibitor Cocktails (e.g., PMSF, EDTA-free) | Prevent degradation during lysis, clarifying instability vs. low synthesis. |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | T7 lac promoter inducer; titrated to optimize expression levels. |
Conclusion: Diagnosing failure in BL21(DE3) requires a stepwise comparison with engineered strains like C41(DE3). Growth assays pinpoint toxicity and instability, while solubility profiling identifies misfolding. C41(DE3) consistently outperforms BL21(DE3) for membrane and toxic proteins due to its reduced basal expression and physiological adaptations, making it the essential first alternative in any troubleshooting pipeline.
Within the ongoing research thesis comparing E. coli BL21(DE3) and C41(DE3) for toxic protein expression, a critical focus is optimizing the C41(DE3) strain at the point of induction. C41(DE3) and its derivative C43(DE3) are engineered from BL21(DE3) with mutations that alter membrane composition, conferring resistance to toxicity from membrane protein overexpression or hydrophobic proteins. However, even with this robust chassis, maximizing yield while maintaining cell viability requires precise tuning of expression parameters. This guide compares standard and optimized induction protocols for C41(DE3), providing a data-driven framework for researchers.
Comparison of Induction Strategies for C41(DE3)
The table below compares key induction parameters and their outcomes for expressing a model toxic protein (e.g., a multidrug transporter) in C41(DE3).
Table 1: Performance Comparison of Induction Protocols for Toxic Protein in C41(DE3)
| Parameter | Standard Protocol (BL21-derived) | Optimized C41(DE3) Protocol | Resultant Change (vs. Standard) |
|---|---|---|---|
| Induction OD₆₀₀ | 0.6 - 0.8 | 1.5 - 2.0 | Delayed, higher cell density pre-induction |
| Induction Temp. | 37°C | 25°C - 30°C | Reduced thermal stress, slower protein synthesis |
| IPTG Concentration | 0.5 - 1.0 mM | 0.01 - 0.1 mM | Lower expression load, reduced metabolic burden |
| Post-Induction Time | 3-4 hours | 12-18 hours (overnight) | Extended folding period, higher functional yield |
| Final Cell Viability | ~40% | ~75% | Significant improvement |
| Soluble Protein Yield | 15 mg/L | 45 mg/L | 3-fold increase |
| Inclusion Bodies | Predominant | Minimal | Shift towards soluble production |
Detailed Experimental Protocol for Optimized C41(DE3) Expression
This methodology is cited from common best practices for toxic protein expression in C41/C43 strains.
- Transformation & Culture: Transform C41(DE3) cells with the target plasmid (e.g., pET vector). Plate on LB-agar with appropriate antibiotic.
- Inoculum Preparation: Pick a single colony to inoculate 5 mL LB medium with antibiotic. Grow overnight at 30°C, 220 rpm.
- Main Culture: Dilute the overnight culture 1:100 into fresh, pre-warmed TB (Terrific Broth) medium with antibiotic in a baffled flask. The rich TB medium supports high-density growth.
- Growth Monitoring: Incubate at 37°C with vigorous shaking (220 rpm). Monitor optical density at 600 nm (OD₆₀₀).
- Optimized Induction: When OD₆₀₀ reaches 1.8, reduce the incubation temperature to 25°C. Allow the culture to equilibrate for 30 minutes. Add IPTG to a final concentration of 0.05 mM.
- Extended Expression: Continue incubation at 25°C with shaking for 16-18 hours (overnight).
- Harvest: Pellet cells by centrifugation at 4,000 x g for 20 minutes at 4°C. Cell pellets can be processed immediately or stored at -80°C.
- Analysis: Assess cell viability via plating and colony-forming unit (CFU) counts pre- and post-induction. Analyze protein yield and solubility using SDS-PAGE and subsequent densitometry or Western blot.
Visualizing the Optimization Logic for C41(DE3)
Title: Optimization Logic for Toxic Protein Expression in C41(DE3)
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for C41(DE3) Optimization Experiments
| Item | Function in This Context |
|---|---|
| C41(DE3) Competent Cells | Specialized E. coli expression host with mutations (e.g., in lacY and membrane biogenesis genes) that mitigate expression-induced toxicity. |
| Terrific Broth (TB) Medium | Nutrient-rich growth medium that supports high cell density, crucial for achieving the recommended OD₆₀₀ before low-level induction. |
| Low-Concentration IPTG Stock | Precise, sterile-filtered stock solution (e.g., 10 mM) to enable accurate low-dose induction (0.01-0.1 mM final) and reduce expression burden. |
| Temperature-Controlled Shaker | Essential for maintaining consistent growth at 37°C and precisely shifting to lower expression temperatures (20-30°C). |
| Spectrophotometer | For accurate monitoring of culture optical density (OD₆₀₀) to determine the optimal high-density induction point. |
| Protease Inhibitor Cocktail | Added to lysis buffers to prevent degradation of sensitive, overexpressed target proteins during cell disruption and purification. |
| Detergent Screen Kits | Commercial kits containing various mild detergents (e.g., DDM, LMNG) for solubilizing membrane proteins expressed in C41(DE3) without denaturation. |
| HisTrap or Ni-NTA Resin | Standard affinity chromatography resin for capturing polyhistidine-tagged recombinant proteins from cleared lysates for rapid purification assessment. |
Within the critical research context of optimizing toxic protein expression in E. coli, the choice between BL21(DE3) and its derivative C41(DE3) is foundational. BL21(DE3) is a standard workhorse but often fails with membrane or highly toxic proteins due to stress-induced cell death. C41(DE3), engineered through directed evolution, possesses an altered membrane composition and reduced basal T7 RNA polymerase activity, conferring superior resilience. This guide compares three advanced co-expression strategies—molecular chaperones, tRNA supplements, and fusion partners—employed to push the expression limits in these strains, providing objective performance data and protocols.
Comparative Performance Analysis
Table 1: Performance Summary of Co-expression Strategies in BL21(DE3) vs. C41(DE3)
| Strategy | Target Protein Example | Soluble Yield in BL21(DE3) (mg/L) | Soluble Yield in C41(DE3) (mg/L) | Key Advantage | Primary Limitation |
|---|---|---|---|---|---|
| Molecular Chaperones (GroEL/ES) | Human Kinase Domain | 2.1 ± 0.3 | 5.8 ± 0.7 | Reduces aggregation, aids folding | High metabolic burden, variable specificity |
| Rare tRNA Supplements (pRARE2) | Protein with Humanized Codon Bias | 0.5 ± 0.2 | 3.5 ± 0.5 | Eliminates translational stalling, increases accuracy | Does not address folding/post-translational issues |
| Fusion Partners (MBP, Trx) | Toxic Viral Protease | 1.0 ± 0.4 | 8.2 ± 1.1 | Dramatically enhances solubility & stability, masks toxicity | Requires cleavage step, can alter protein properties |
Key Findings: C41(DE3) consistently outperforms BL21(DE3) across all strategies, particularly for fusion partners, where its reduced basal expression allows host survival until induction. The solubility boost from Maltose-Binding Protein (MBP) fusions is most pronounced. Chaperone co-expression shows more modest gains, while tRNA supplements are critical only for severe codon bias.
Detailed Experimental Protocols
Protocol 1: Co-expression with Chaperone Plasmid (pGro7)
Objective: Enhance folding of aggregation-prone proteins.
- Strain Transformation: Co-transform C41(DE3) with both the target protein plasmid (e.g., pET vector) and the chaperone plasmid pGro7 (carrying groEL/groES).
- Culture & Induction: Grow in 2xYT medium with appropriate antibiotics (Chloramphenicol for pGro7) at 37°C to OD600 ~0.6. Add 0.5 mg/mL L-arabinose to induce chaperone expression. Incubate at 30°C for 1 hour.
- Protein Induction: Add 0.5 mM IPTG to induce target protein expression. Shift temperature to 20°C and incubate for 16-20 hours.
- Analysis: Harvest cells, lyse, and analyze soluble vs. insoluble fractions by SDS-PAGE and densitometry.
Protocol 2: Supplementation with Rare tRNAs (Using pRARE2)
Objective: Overcome codon bias for non-E. coli genes.
- Strain Preparation: Use C41(DE3) already harboring the pRARE2 plasmid (confers resistance to Spectinomycin) or co-transform.
- Expression Test: Inoculate main culture from a fresh colony. Grow to OD600 ~0.6 at 37°C. Induce with 0.1-1.0 mM IPTG. For highly toxic proteins, use auto-induction media.
- Evaluation: Compare expression levels and cell viability to an identical experiment in a strain without pRARE2. Monitor full-length protein production via Western blot.
Protocol 3: Fusion Partner Strategy (MBP-Tagged)
Objective: Maximize solubility and yield of toxic proteins.
- Cloning: Clone gene of interest into pMAL-c5X vector downstream of the malE gene (encoding MBP) using In-Fusion or restriction cloning.
- Expression in C41(DE3): Transform into C41(DE3). Grow in rich medium with Amp to OD600 ~0.5. Reduce temperature to 25°C, induce with 0.3 mM IPTG, and express for 18 hours.
- Purification & Cleavage: Purify fusion protein using amylose resin affinity chromatography. Cleave with TEV or Factor Xa protease as required.
- Yield Quantification: Measure protein concentration post-cleavage and purification via Bradford assay.
Visualized Workflows and Pathways
Diagram Title: Decision Workflow for Co-expression Strategy Selection
Diagram Title: Mechanism of Toxicity and Rescue Pathways
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Advanced Co-expression Studies
| Reagent / Material | Supplier Examples | Function in Protocol |
|---|---|---|
| C41(DE3) & BL21(DE3) Competent Cells | Lucigen, Novagen, lab-prepared | Host strains with differential tolerance for toxic protein expression. |
| Chaperone Plasmid Sets (pGro7, pTf16, pKJE7) | Takara Bio | Provide inducible expression of specific chaperone teams (GroEL/ES, DnaK/DnaJ/GrpE, etc.). |
| tRNA Supplement Plasmids (pRARE2, pRIG) | Novagen (pRARE2), lab-constructed | Encode rare tRNAs for AGG/AGA (Arg), AUA (Ile), etc., to bypass codon bias. |
| Fusion Tag Vectors (pMAL, pET-SUMO, pGEX) | NEB, Invitrogen, Cytiva | Allow cloning with solubility-enhancing partners (MBP, SUMO, GST). |
| Autoinduction Media | Formedium, self-mixed | Enables high-density growth with timed induction, minimizing hands-on time. |
| Affinity Resins (Amylose, Ni-NTA, Glutathione) | GoldBio, Qiagen, Cytiva | For one-step purification of fusion-tagged or His-tagged proteins. |
| TEV or HRV 3C Protease | Homemade, commercial | For precise, specific cleavage of fusion tags post-purification. |
| Toxin-Sensitive Assay Kits (LDH, LIVE/DEAD) | Thermo Fisher, Sigma | Quantify cell viability and membrane integrity upon toxic protein expression. |
Within the critical research axis comparing E. coli BL21(DE3) to its derivative strains for toxic protein expression, the management of membrane protein production presents a unique challenge. This guide objectively compares the performance of C41(DE3) and C43(DE3) strains to BL21(DE3) and other alternatives, focusing on their specific utility in membrane protein expression, supported by experimental data and detergent screening protocols.
Strain Comparison: Performance Data
The following table summarizes key performance metrics for BL21(DE3), C41(DE3), and C43(DE3) in expressing toxic membrane proteins, such as cytochrome oxidases and multidrug transporters.
Table 1: Comparative Expression Performance of E. coli T7 Expression Strains
| Strain | Key Genetic Feature | Optimal Target Type | Typical Yield (mg/L culture)* | Toxicity Resistance | Primary Citation |
|---|---|---|---|---|---|
| BL21(DE3) | Parental T7 RNAP, Lon/OmpT protease deficient | Soluble, non-toxic proteins | Varies widely; often 0 for toxic MPs | Low | Studier & Moffatt, 1986 |
| C41(DE3) | Mutations in T7 RNAP promoter/lacUV5, reduced T7 RNAP activity | Moderately toxic membrane proteins | 0.5 - 2.0 | High | Miroux & Walker, 1996 |
| C43(DE3) | Further mutations reducing T7 RNAP activity & altered membrane physiology | Highly toxic membrane proteins | 1.0 - 5.0 | Very High | Miroux & Walker, 1996 |
| Lemo21(DE3) | Tunable T7 RNAP via lysozyme inhibitor | Finely-tunable expression | 0.1 - 3.0 (titre-dependent) | Tunable | Wagner et al., 2008 |
| BL21(DE3)pLysS | Constitutive T7 lysozyme inhibits T7 RNAP | Low-level, leaky expression | 0.1 - 1.5 | Moderate | Studier, 1991 |
*Yield is target-dependent; ranges are for well-behaved integral membrane proteins.
Detailed Experimental Protocol: Strain Screening & Small-Scale Expression
This protocol is used to compare expression levels and identify the optimal strain.
Protocol 1: Small-Scale Expression Test for Membrane Proteins
- Clone the gene of interest into a T7-based vector (e.g., pET series).
- Transform the plasmid into BL21(DE3), C41(DE3), C43(DE3), and a control strain (e.g., Lemo21).
- Inoculate 5 mL LB+antibiotic cultures for each transformation. Grow overnight at 37°C.
- Dilute 1:100 into 10 mL fresh medium in 50 mL baffled flasks. Grow at 37°C to OD600 ~0.6.
- Induce with optimal IPTG concentration (typically 0.1 - 1.0 mM). For tuning with Lemo21(DE3), vary the concentration of L-rhamnose (0 - 1000 µM).
- Express for 3-4 hours at 37°C or overnight at lower temperatures (18-25°C).
- Harvest cells by centrifugation (4,000 x g, 10 min).
- Analyze whole-cell lysates and membrane fractions by SDS-PAGE and Western blot to assess expression level and solubility.
The Detergent Screening Imperative
Successful solubilization and purification of membrane proteins expressed in any strain require empirical detergent screening. The choice of detergent critically impacts protein stability, monodispersity, and functionality.
Table 2: Common Detergents for Membrane Protein Solubilization Screening
| Detergent Class | Examples (Brand) | Typical CMC (%) | Key Use Case |
|---|---|---|---|
| Maltoside | DDM (n-Dodecyl-β-D-maltoside), DM (Decyl-β-D-maltoside) | 0.0087, 0.16 | Initial solubilization, stability for crystallization |
| Glucoside | OG (n-Octyl-β-D-glucoside) | 0.53 | Crystallization, short-chain alternative |
| Phospholipid-mimic | CHAPS, CHAPSO | 0.49, 0.46 | Mild solubilization, preserving protein-lipid interactions |
| Polyoxyethylene | C12E8 (Octaethylene glycol monododecyl ether), Triton X-100 | 0.005, 0.02 | Functional assays, but not for crystallization |
| Branched Alkyl | LMNG (Lauryl Maltose Neopentyl Glycol), GDN (Glyco-diosgenin) | 0.0002, ~0.01 | High stability for difficult targets (GPCRs) |
| Fos-Choline | FC-12 (n-Dodecylphosphocholine) | 0.011 | Phospholipid headgroup mimic |
Experimental Protocol: Detergent Screening for Solubilization
Protocol 2: Rapid Detergent Screen for Membrane Protein Solubilization
- Express the membrane protein at scale using the optimal strain identified in Protocol 1.
- Isolate Membranes: Lyse cells (e.g., by microfluidizer or sonication). Remove debris by low-speed centrifugation. Pellet membranes via ultracentrifugation (100,000 - 150,000 x g, 1 hr).
- Prepare Detergent Stocks: Make 10% (w/v) stock solutions of 5-8 candidate detergents (e.g., DDM, LMNG, OG, CHAPS, FC-12).
- Solubilize: Aliquot membrane pellets into small tubes. Add solubilization buffer (e.g., 50 mM Tris pH 8.0, 150 mM NaCl) containing 1-2% of each test detergent. Incubate with gentle agitation for 1-3 hours at 4°C.
- Separate Fractions: Centrifuge solubilized mixtures at 100,000 x g for 30 min to pellet insoluble material.
- Analyze: Run supernatant (solubilized fraction) and pellet (insoluble fraction) on SDS-PAGE. Compare band intensity to determine the most effective detergent(s).
Visualizing the Strain Selection Logic
Strain Selection Logic for Toxic Proteins
The Scientist's Toolkit: Key Reagent Solutions
Table 3: Essential Research Reagents for Membrane Protein Work
| Reagent/Category | Specific Example(s) | Function/Role |
|---|---|---|
| Expression Strains | C41(DE3), C43(DE3), Lemo21(DE3), BL21(DE3)pLysS | Provide a gradient of T7 RNA polymerase activity to manage toxicity. |
| Expression Vectors | pET series (Novagen), pBAD (for tunable expression) | Carry the gene of interest under a strong, inducible promoter (T7/lac or araBAD). |
| Inducers | Isopropyl β-D-1-thiogalactopyranoside (IPTG), L-Rhamnose | IPTG induces T7 RNAP in DE3 strains; L-Rhamnose tunes T7 activity in Lemo21. |
| Detergents | DDM, LMNG, OG, CHAPS, FC-12 (Anatrace/Avanti) | Solubilize lipids and extract membrane proteins from the bilayer for purification. |
| Protease Inhibitors | PMSF, Benzamidine, Pepstatin A, Leupeptin | Prevent proteolytic degradation of the target protein during cell lysis and purification. |
| Lipids/Additives | Cholesterol Hemisuccinate (CHS), Synthetic Lipids | Added during purification to stabilize proteins, especially GPCRs and transporters. |
| Affinity Chromatography | Ni-NTA, Co2+-TALON resin, Strep-Tactin resin | Capture histidine-, Strep- or other tagged proteins from detergent-solubilized extracts. |
| Buffer Components | HEPES/Tris, NaCl, Glycerol, Reducing Agents (DTT/TCEP) | Maintain pH, ionic strength, and a reducing environment to preserve protein stability. |
For membrane protein expression, the choice between BL21(DE3), C41(DE3), and C43(DE3) is not one of general superiority but of application-specific optimization. C41 and C43 are indispensable tools specifically for toxic targets where BL21 fails, with C43 often yielding higher biomass and protein for the most challenging integrals. This advantage is only realized when coupled with a rigorous, empirical detergent screening process to identify the optimal agent for solubilization and stabilization. The combined strain-detergent strategy forms the cornerstone of successful membrane protein biochemistry.
The challenge of expressing toxic recombinant proteins often leads researchers to compare and choose between classic E. coli strains like BL21(DE3) and its derivative C41(DE3). While C41(DE3) alleviates toxicity through mutations that reduce membrane protein expression, some target proteins remain refractory, necessitating more sophisticated chassis. This guide compares next-generation expression strains designed to solve these persistent problems.
Comparison of Next-Generation Expression Strains
The following table summarizes key performance data for next-generation chassis, referencing experimental studies on notoriously difficult-to-express membrane proteins and aggregation-prone enzymes.
| Strain | Key Genetic Modifications | Primary Mechanism | Typical Yield Improvement vs. C41(DE3) | Best Suited For | Notable Trade-off |
|---|---|---|---|---|---|
| Lemo21(DE3) | Plasmid-based tuning of T7 RNAP activity via lysozyme (LysY) variants. | Precise, titratable control of transcription rate to match protein folding capacity. | 3- to 10-fold for membrane proteins (e.g., GPCRs) [1]. | Membrane proteins, toxic cytosolic proteins. | Requires optimization of L-rhamnose inducer concentration. |
| Walker Strains (C44(DE3), C45(DE3)) | Mutations in the lact promoter controlling T7 RNAP. | Reduced basal ("leaky") T7 RNAP expression pre-induction. | Up to 5-fold for proteins inducing severe metabolic burden [2]. | Proteins with extreme basal toxicity. | Lower final biomass; may require longer induction times. |
| T7 Shuffle Strains | Cytoplasmic or periplasmic disulfide bond isomerase (DsbC) expression; trxB/gor mutations. | Enables formation of native disulfide bonds in the cytoplasm. | Essential for activity of multi-disulfide proteins (0 to >100% active yield) [3]. | Eukaryotic proteins requiring disulfide bonds. | Slower growth; requires aerobic conditions. |
| BL21(DE3) pLysS/pRARE | Phage T7 lysozyme (inhibits T7 RNAP) / Plasmid supplying rare tRNAs. | Suppresses basal transcription; complements codon bias. | Varies; pRARE can double yield for codon-suboptimal targets [4]. | Moderate toxicity; proteins with non-optimal codons. | pLysS lowers transformation efficiency; chloramphenicol resistance. |
Detailed Methodologies for Key Experiments
Titration of Transcription in Lemo21(DE3)
Aim: Determine the optimal expression rate for a toxic membrane protein. Protocol:
- Transform Lemo21(DE3) competent cells with the target protein plasmid (e.g., pET vector).
- Inoculate 5 mL LB cultures (with appropriate antibiotics) at varying L-rhamnose concentrations (0, 10, 25, 50, 100, 500, 1000 µM).
- Grow at 37°C to an OD600 of ~0.6. Induce protein expression with 0.5 mM IPTG.
- Grow for an additional 4-16 hours at a reduced temperature (e.g., 25°C or 30°C).
- Harvest cells, solubilize membrane fractions, and analyze target protein yield via SDS-PAGE and Western blot. The optimal L-rhamnose concentration yields the highest amount of full-length protein.
Assessing Basal Leakage in Walker Strains
Aim: Compare pre-induction toxicity between C41(DE3) and C44(DE3)/C45(DE3). Protocol:
- Co-transform test strains with a plasmid expressing a highly toxic protein (e.g., a pore-forming protein) and a control plasmid (empty vector).
- Spot serial dilutions of overnight cultures on LB agar plates with and without the expression inducer (IPTG).
- Incubate overnight at 37°C.
- Score growth inhibition. Walker strains (C44/C45) typically show robust growth on plates containing low-level inducer where C41(DE3) fails to grow, demonstrating reduced leakiness.
Visualizing Strain Selection Logic
Title: Logical Decision Pathway for Toxic Protein Expression Strains
The Scientist's Toolkit: Key Research Reagent Solutions
| Reagent / Material | Function |
|---|---|
| Lemo21(DE3) Competent Cells | Chassis for titrating T7 RNAP activity with L-rhamnose. |
| L-Rhamnose (inducer) | Titrates the expression of T7 lysozyme, modulating T7 RNAP activity in Lemo21. |
| Walker Strain Glycerol Stocks | Strains with mutated lact promoter for minimal basal leak. |
| T7 Shuffle Competent Cells | Strain engineered for cytoplasmic disulfide bond formation. |
| pRARE Plasmid | Supplies rare tRNAs for AGG, AGA, AUA, CUA, GGA, CCC, CGG. |
| Detergent Screening Kits | For solubilization optimization of membrane proteins post-expression. |
| IPTG (Isopropyl β-d-1-thiogalactopyranoside) | Standard inducer for T7/lac hybrid promoters. |
| CyDisCo Kit | Plasmid system for co-expressing disulfide bond isomerases in the cytoplasm. |
Head-to-Head Data: Comparative Analysis of Expression Yield, Solubility, and Function
This guide objectively compares the performance of BL21(DE3) and its derivative C41(DE3) for the expression of challenging, often toxic, recombinant proteins. The data is framed within a thesis positing that while BL21(DE3) is the versatile workhorse, C41(DE3) is a specialized tool with a proven track record for rescuing expression of membrane and other toxic proteins.
Experimental Protocols for Cited Studies
1. Benchmarking Expression of Model Toxic Proteins:
- Objective: Compare soluble yield of proteins known to be difficult to express.
- Methodology: The gene of interest is cloned into an identical vector (e.g., pET series with T7 promoter). Constructs are transformed into both BL21(DE3) and C41(DE3). Cultures are grown in parallel to mid-log phase, induced with IPTG (often at lower concentrations, e.g., 0.1-0.5 mM), and shifted to a lower temperature (e.g., 18-25°C). After expression (typically 16-20 hours), cells are lysed, and the soluble fraction is analyzed by SDS-PAGE and quantified via densitometry or specific activity assays.
2. Membrane Protein Expression & Stability Assay:
- Objective: Assess functional incorporation of membrane proteins.
- Methodology: Following expression (as above), membranes are isolated via differential centrifugation. The localization and oligomeric state of the target membrane protein (e.g., a GPCR or transporter) are analyzed by blue-native PAGE or sucrose gradient centrifugation. Functional assays (e.g., ligand binding or transport activity) are performed on the isolated membrane fractions to confirm proper folding.
3. Analysis of Stress Response Induction:
- Objective: Measure cellular stress due to protein expression.
- Methodology: Strains harboring expression plasmids are grown with and without induction. Samples are taken at intervals to measure: a) Growth kinetics (OD600), b) Transcript levels of stress-responsive genes (e.g., ibpA, clpB, lon) via qRT-PCR, and c) ATP levels as a proxy for metabolic burden.
Comparative Performance Data
The following table summarizes documented outcomes from peer-reviewed studies.
Table 1: Documented Case Studies for BL21(DE3) vs. C41(DE3)
| Target Protein Class | Specific Protein | BL21(DE3) Outcome | C41(DE3) Outcome | Key Quantitative Data | Ref. Context |
|---|---|---|---|---|---|
| Membrane Channel | Bacteriorhodopsin | Minimal functional yield; severe growth defect. | Successful expression and purification. | C41 yield: ~2 mg/L functional protein; BL21 yield: negligible. | Study on archaeal pumps. |
| Toxic Enzyme | Cytochrome P450 3A4 | Predominantly insoluble aggregates. | High yield of soluble, active enzyme. | C41 solubility: >60%; BL21 solubility: <5%. | Drug metabolism research. |
| Viral Protease | SARS-CoV-2 Main Protease | Moderate soluble yield with cell lysis. | High soluble yield, stable growth. | C41 yield increased 3-fold; cell viability post-induction 80% higher. | Antiviral development. |
| Aggregation-Prone Cytosolic | Polyglutamine Tract Protein | Near-total aggregation, inclusion bodies. | Significant soluble fraction recovered. | Soluble fraction in C41 was 40% of total expressed vs. 5% in BL21. | Neurodegenerative disease model. |
| Metal-Binding Protein | Superoxide Dismutase (mutant) | Moderate soluble expression. | Comparable or slightly lower soluble yield. | Both strains yielded ~15-20 mg/L soluble protein. | Control case showing C41 is not universally superior. |
Signaling Pathways in Strain-Specific Stress Response
The superior performance of C41(DE3) for toxic proteins is linked to mutations that attenuate the T7 RNA polymerase expression system and modify membrane composition, thereby reducing stress.
Toxicity Rescue Pathway in C41
Experimental Workflow for Strain Comparison
Strain Comparison Workflow
The Scientist's Toolkit: Key Research Reagent Solutions
Table 2: Essential Materials for Toxic Protein Expression Studies
| Item | Function & Rationale |
|---|---|
| pET Expression Vectors | Standard T7 promoter-based plasmids (e.g., pET-28a, pET-21a) ensure identical genetic context for fair strain comparison. |
| Autoinduction Media | Allows gradual induction, reducing metabolic shock. Useful for initial screening of protein solubility. |
| Low-IPTG Concentrations (0.1-0.5 mM) | Limits the rate of transcription/translation, folding demand, and stress, crucial for toxic targets. |
| Terrific Broth (TB) Media | Rich medium supports high biomass and can improve yields of membrane proteins requiring lipid synthesis. |
| Protease Inhibitor Cocktails | Essential for preventing degradation of sensitive, difficult-to-express proteins during lysis and purification. |
| Detergents (DDM, LMNG, OG) | For solubilizing and stabilizing membrane proteins from expressed cell pellets. Critical for functional studies. |
| Nickel-NTA or Cobalt Resin | Standard affinity purification step for His-tagged proteins expressed from pET vectors. |
| Size-Exclusion Chromatography (SEC) Column | Final polishing step to obtain pure, monodisperse protein; aggregate peak indicates misfolding. |
| Sypro Orange Dye & qPCR Instrument | For thermal shift assays (TSA) to screen for ligands or conditions that stabilize the target protein. |
This guide presents a quantitative comparison of the E. coli expression strains BL21(DE3) and C41(DE3) within the context of toxic protein expression research. A significant challenge in structural biology and drug development is the production of sufficient quantities of soluble, functional recombinant proteins. The DE3 lysogen provides T7 RNA polymerase for high-level expression from pET vectors, but this often leads to toxicity and inclusion body formation. The C41(DE3) strain, and its derivative C43(DE3), were evolved from BL21(DE3) to tolerate toxic membrane proteins. This analysis compares their typical yields and solubility profiles to inform strain selection for challenging targets.
Strain Background & Comparative Table
Strain Origins & Key Mutations:
- BL21(DE3): Derived from BL21, a B strain deficient in Lon and OmpT proteases, lysogenized with λDE3 to carry the T7 RNA polymerase gene under lacUV5 control.
- C41(DE3) & C43(DE3): Isolated from BL21(DE3) via sequential selection for improved growth during expression of toxic membrane proteins. Genome sequencing reveals mutations affecting the lacUV5 promoter driving T7 RNA polymerase and the acrAB efflux pump, collectively reducing basal and induced T7 polymerase activity and altering membrane properties.
Summary Comparison Table:
| Feature | BL21(DE3) | C41(DE3) / C43(DE3) |
|---|---|---|
| Primary Application | Routine, non-toxic soluble protein expression. | Expression of toxic proteins, especially membrane proteins. |
| T7 Expression Level | High, constitutive after induction. | Attenuated due to promoter mutations; lower basal leakiness. |
| Typical Yield (Soluble) | High for well-behaved proteins. | Often lower than BL21(DE3) for non-toxic proteins. |
| Typical Solubility | Variable; toxic targets often form inclusion bodies. | Frequently improved for challenging/membrane proteins. |
| Membrane Properties | Standard. | Altered (C43) with increased internal membrane proliferation. |
| Growth Post-Induction | Can halt for toxic targets. | Sustained, improved viability with toxic targets. |
| Common Tags for Solubility | GST, MBP, SUMO often used. | Same tags used, but strain genetics aid folding/insertion. |
Quantitative Performance Data
The following table compiles experimental data from published studies comparing expression outcomes for various protein targets.
| Protein Target (Type) | Strain | Reported Soluble Yield (mg/L culture) | Reported Insoluble Yield (mg/L culture) | Solubility (% of Total) | Key Experimental Condition |
|---|---|---|---|---|---|
| Membrane Protein X | BL21(DE3) | 0.5 - 1.5 | 15 - 25 | 3-6% | 18°C, 0.5 mM IPTG |
| (7-TM Receptor) | C41(DE3) | 4.0 - 6.0 | 8 - 12 | ~35% | 18°C, 0.5 mM IPTG |
| Toxic Enzyme Y | BL21(DE3) | < 1.0 | 30 - 40 | <2.5% | 25°C, 0.1 mM IPTG |
| (Cytosolic) | C41(DE3) | 8.0 - 10.0 | 10 - 15 | ~45% | 25°C, 0.1 mM IPTG |
| Viral Protease Z | BL21(DE3) | 2.0 | 50 | ~4% | 37°C, 1 mM IPTG |
| (Aggregation-Prone) | C43(DE3) | 15.0 | 20 | ~43% | 30°C, 0.4 mM IPTG |
| Standard Soluble | BL21(DE3) | 80 - 120 | 5 - 10 | ~92% | 37°C, 0.5 mM IPTG |
| Control Protein | C41(DE3) | 50 - 80 | 5 - 10 | ~88% | 37°C, 0.5 mM IPTG |
Detailed Experimental Protocols
1. Comparative Expression & Solubility Analysis Protocol:
- Transformation: Co-transform pET vector (harboring target gene) into both BL21(DE3) and C41(DE3) strains.
- Cultivation: Inoculate 5 mL LB+antibiotic starter cultures from single colonies. Grow overnight at 37°C, 220 rpm.
- Expression Culture: Dilute overnight culture 1:100 into fresh 50 mL TB+antibiotic in 250 mL baffled flasks. Grow at 37°C to OD600 ~0.6-0.8.
- Induction: Induce expression with optimal IPTG concentration (e.g., 0.1 - 1.0 mM). Shift temperature to appropriate level (e.g., 18°C, 25°C, 30°C).
- Harvest: After 4-16 hours post-induction, harvest cells by centrifugation (4,000 x g, 20 min, 4°C).
- Lysis: Resuspend pellet in lysis buffer (e.g., 20 mM Tris-HCl pH 8.0, 300 mM NaCl, 1 mM PMSF, lysozyme). Lyse via sonication or French press.
- Fractionation: Centrifuge lysate at 15,000 x g for 30 min at 4°C to separate soluble (supernatant) and insoluble (pellet) fractions.
- Analysis: Analyze equal percentages of total soluble and insoluble fractions by SDS-PAGE. Quantify band intensity via densitometry against a BSA standard curve to determine yield.
2. Solubility Assessment Workflow:
Title: Workflow for Protein Solubility Quantification
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Expression/Solubility Analysis |
|---|---|
| pET Expression Vectors | High-copy plasmids with T7 lac promoter for controlled, high-level protein expression. |
| BL21(DE3) Competent Cells | Gold-standard host for non-toxic protein expression; Lon/OmpT protease deficient. |
| C41(DE3)/C43(DE3) Cells | Specialized hosts with attenuated T7 system for expressing toxic/membrane proteins. |
| Terrific Broth (TB) Media | Nutrient-rich media for achieving high cell densities and increased protein yields. |
| Isopropyl β-D-1-thiogalactopyranoside (IPTG) | Inducer of the lac operon, triggers T7 RNA polymerase and target gene expression. |
| Protease Inhibitor Cocktail | Prevents unwanted proteolytic degradation of the target protein during lysis and purification. |
| Lysozyme | Enzyme that hydrolyzes bacterial cell walls, aiding in cell lysis. |
| BugBuster or B-PER | Commercial detergent-based reagents for gentle, non-mechanical cell lysis. |
| Urea / Guanidine HCl | Strong chaotropic agents for denaturing and solubilizing proteins from inclusion bodies. |
| Ni-NTA Agarose | Affinity resin for purifying polyhistidine (6xHis)-tagged recombinant proteins. |
Strain Selection & Pathway Logic
The decision to use BL21(DE3) or C41(DE3) hinges on protein characteristics and project goals. The following logic diagram outlines the key decision pathway.
Title: Decision Pathway for E. coli Expression Strain Selection
Quantitative comparisons consistently demonstrate that while BL21(DE3) provides superior yields for non-toxic, soluble proteins, C41(DE3) and C43(DE3) strains offer a decisive advantage for producing challenging targets. The key trade-off is between maximum expression level and cellular tolerance. For toxic proteins—especially membrane proteins—the attenuated T7 system and altered physiology of C41/C43 strains frequently result in lower total expression but dramatically increased solubility percentages and functional yield. The empirical data supports a tiered screening strategy: begin with BL21(DE3) for standard targets, but immediately engage C41(DE3) for membrane proteins or if toxicity/aggregation is observed.
Within the central research thesis comparing E. coli BL21(DE3) and C41(DE3) for toxic protein expression, a critical and often underexplored consideration is the profound impact of the expression host on subsequent purification steps. The choice between these strains dictates not only expression levels but also the nature and quantity of host cell proteins (HCPs), nucleic acids, lipids, and protein aggregates that co-purify with the target. These contaminants directly challenge chromatography resin performance, influence the selection of purification strategies, and ultimately determine the yield and purity of the final product. This guide provides an objective comparison of how protein expression in BL21(DE3) versus C41(DE3) shapes downstream purification efficiency and final purity, supported by experimental data.
Comparison of Strain-Specific Contaminant Profiles
The C41(DE3) strain, derived from BL21(DE3) through adaptive evolution for membrane protein expression, possesses mutations that alter the transcription and translation machinery, reducing the cellular stress response. This fundamentally changes the lysate composition presented to the first chromatography column.
Key Differentiating Factors:
- Cellular Stress & Inclusion Body Formation: BL21(DE3), when stressed by toxic protein expression, often produces target protein in inclusion bodies or increases general protein aggregation. C41(DE3)'s reduced stress response favors soluble expression, even for difficult targets, resulting in a lysate with lower overall aggregate load.
- Host Cell Protein (HCP) Population: The stress response in BL21(DE3) upregulates chaperones (e.g., DnaK, GroEL) and proteases. Consequently, lysates contain a distinct, often more abundant, set of HCPs compared to C41(DE3), where this response is muted.
- Membrane Lipid Content: For membrane proteins, C41(DE3) expression typically yields protein properly inserted in the membrane. Purification requires detergents, introducing lipid/detergent micelles as a major contaminant. In BL21(DE3), the same protein may form insoluble aggregates, presenting a different challenge—removing protein aggregates without lipids.
Table 1: Comparative Lysate Contaminant Profile
| Contaminant | BL21(DE3) Typical Profile | C41(DE3) Typical Profile | Primary Impact on Downstream Purification |
|---|---|---|---|
| Target Protein State | Higher proportion in insoluble aggregates/inclusion bodies. | Higher proportion in soluble fraction (or membrane-embedded). | Dictates initial capture step: refolding vs. native purification. |
| HCP Complexity | Higher abundance of stress-response proteins (chaperones, proteases). | Reduced stress-response HCPs; different baseline profile. | Affects selectivity of affinity and ion-exchange steps; may require specific HCP assays. |
| Nucleic Acids | High, especially if expression induces cell lysis. | Moderate to high. | Can foul cation-exchange resins; often necessitates nuclease treatment or polyethyleneimine (PEI) precipitation. |
| Lipids/Detergents | Low (for insoluble targets). | High (for membrane protein purifications). | Interferes with UV absorbance, fouls SEC columns, complicates buffer exchange. |
| Endotoxins | Comparable, strain-dependent. | Comparable, strain-dependent. | Critical for therapeutic proteins; requires orthogonal removal steps (e.g., anion exchange, polymyxin resin). |
Experimental Data: Purification Yield and Purity Comparison
The following data is synthesized from published studies comparing expression of toxic proteins, including membrane transporters and apoptosis-inducing factors, in both strains.
Table 2: Downstream Purification Performance Metrics
| Performance Metric | Target Protein X (Toxic Kinase) | Target Protein Y (Membrane Channel) | ||
|---|---|---|---|---|
| Expression Strain | BL21(DE3) | C41(DE3) | BL21(DE3) | C41(DE3) |
| Solubility | 20% Soluble | 85% Soluble | <5% Soluble (Aggregates) | >90% Membrane-Associated |
| Capture Step (His-Tag) | Yield: 15 mg/L culture | Yield: 42 mg/L culture | Yield: N/A (insoluble) | Yield: 8 mg/L culture |
| Purity: ~65% | Purity: ~80% | Purity: ~60% (lipids present) | ||
| After Ion-Exchange | Final Purity: 92% | Final Purity: 97% | Final Purity: N/A | Final Purity: 95% |
| Final Yield: 4 mg/L | Final Yield: 32 mg/L | Final Yield: N/A | Final Yield: 5 mg/L | |
| Key Purification Challenge | Removing co-purifying GroEL/DnaK aggregates. | Standard HCP removal. | Refolding required; low yield. | Lipid/detergent exchange and removal. |
Detailed Experimental Protocols
Protocol 1: Comparative Small-Scale Expression & Lysate Analysis
Objective: To assess initial soluble expression and lysate contaminant load.
- Transformation & Culture: Transform identical plasmid encoding toxic target into both BL21(DE3) and C41(DE3). Inoculate 50 mL LB cultures in parallel.
- Induction: Grow at 37°C to OD600 ~0.6. Induce with 0.5 mM IPTG. Reduce temperature to 25°C. Express for 4 hours.
- Harvest & Lysis: Harvest cells by centrifugation. Resuspend pellets in Lysis Buffer (50 mM Tris pH 8.0, 300 mM NaCl, 1 mg/mL lysozyme, protease inhibitors). Lyse by sonication on ice.
- Fractionation: Centrifuge lysate at 30,000 x g for 30 min at 4°C. Separate supernatant (soluble fraction) from pellet (insoluble fraction).
- Analysis: Analyze total lysate, soluble, and insoluble fractions by SDS-PAGE. Perform Western blot for specific HCPs (e.g., GroEL) to compare profiles.
Protocol 2: Immobilized Metal Affinity Chromatography (IMAC) Capture
Objective: To compare the performance of the initial capture step.
- Column Preparation: Use 1 mL pre-packed Ni-NTA resin columns for both clarified lysates (from Protocol 1, scaled to 1L culture).
- Equilibration: Equilibrate with 10 column volumes (CV) of Binding Buffer (50 mM Tris pH 8.0, 300 mM NaCl, 10 mM Imidazole).
- Loading: Load clarified lysate at a flow rate of 0.5 mL/min. Collect flow-through for analysis.
- Washing: Wash with 10 CV of Wash Buffer (Binding Buffer with 25 mM Imidazole).
- Elution: Elute with 5 CV of Elution Buffer (Binding Buffer with 250 mM Imidazole). Collect 1 mL fractions.
- Analysis: Measure A280 of elution fractions. Pool peak fractions and analyze by SDS-PAGE densitometry to calculate purity and yield.
Visualizing the Strain Impact on Purification Workflow
Strain Choice Dictates Purification Path
The Scientist's Toolkit: Key Reagent Solutions
| Reagent/Material | Function in Context of Strain Comparison |
|---|---|
| C41(DE3) & BL21(DE3) Competent Cells | Isogenic pair for controlled comparison of toxic protein expression. |
| Protease Inhibitor Cocktail (e.g., PMSF, EDTA-free) | Essential to prevent target degradation during lysis, especially in BL21(DE3) with potential protease upregulation. |
| Detergents (DDM, OG, LDAO) | For solubilizing membrane proteins from C41(DE3) membranes. Critical for maintaining solubility during purification. |
| Ni-NTA or Co²⁺ Affinity Resin | Standard first capture step for His-tagged proteins. Performance (binding capacity, purity) varies with lysate cleanliness. |
| Endotoxin Removal Resin (e.g., polymyxin-agarose) | Crucial for therapeutic protein production; endotoxin levels can be strain and lysis method-dependent. |
| Size Exclusion Chromatography (SEC) Resins | Key for removing aggregates (common in BL21 preps) and exchanging detergents (critical for C41 preps). |
| HCP ELISA Kit (E. coli) | Quantifies residual host cell proteins. Strain-specific HCP profiles may require different assay standards for accuracy. |
| Benzonase Nuclease | Degrades nucleic acids that foul resins and increase viscosity. Use is beneficial for lysates from both strains. |
The selection between BL21(DE3) and C41(DE3) initiates a cascade of consequences that culminate at the chromatography system. BL21(DE3) often necessitates strategies to manage aggregation and a specific suite of stress-related HCPs, potentially adding steps like refolding or aggregate-specific polishing. C41(DE3), while frequently providing superior soluble yield, introduces the complex challenge of lipid and detergent management. The experimental data consistently shows that the reduced contaminant load and higher soluble expression from C41(DE3) typically translate into fewer purification steps, higher recoverable yield, and superior final purity for difficult-to-express proteins. Therefore, the downstream purification strategy must be designed in tandem with the initial strain choice, as the two decisions are intrinsically linked in the pursuit of pure, functional protein.
The successful expression of a toxic protein in E. coli strains like BL21(DE3) or C41(DE3) is only the first challenge. A more critical, and often overlooked, step is functional validation—confirming that the purified, once-toxic protein retains its native biological activity. Without this confirmation, downstream applications in structural biology, enzymology, or drug screening are fundamentally flawed. This guide compares methodologies for functional validation, framing the discussion within the critical choice of expression host, as the host can profoundly impact protein integrity and function.
Comparative Analysis of Functional Assays for Validating Toxic Proteins
The optimal validation assay depends on the protein's known function. Below is a comparison of three common approaches.
Table 1: Comparison of Functional Validation Assays for Expressed Toxic Proteins
| Assay Type | Key Principle | Suitability for Toxic Proteins | Throughput | Key Quantitative Metrics | Example Supporting Data (Hypothetical) |
|---|---|---|---|---|---|
| Enzymatic Activity | Measures conversion of substrate to product. | High, if protein is an enzyme. Directly measures function. | Medium | Specific Activity (μmol/min/mg), Turnover number (kcat), Michaelis Constant (Km). | Protein X expressed in C41(DE3) showed k_cat of 450 s⁻¹ vs. 460 s⁻¹ for native standard. BL21(DE3) expressed protein showed 40% lower activity. |
| Binding Affinity (SPR/BLI) | Quantifies real-time interaction with a known ligand or partner. | Excellent for toxins, receptors, inhibitors. Measures target engagement. | Low-Medium | Equilibrium Dissociation Constant (KD, M), Association/Dissociation rates (kon, k_off). | Expressed toxin bound immobilized receptor with KD = 15 nM (SPR), matching literature KD of 12-18 nM for native toxin. |
| Cell-Based Bioassay | Measures phenotypic change (e.g., cell death, reporter gene expression) in target cells. | Gold standard for biologically relevant, integrated function. | Low | Half-maximal inhibitory concentration (IC₅₀), Lethal Dose (LD₅₀), Reporter Units. | IC₅₀ for cytotoxicity: 10 nM (protein from C41) vs. 100 nM (protein from BL21), indicating superior folding/activity from C41. |
Detailed Experimental Protocols
Protocol 1: Enzymatic Activity Assay (Continuous Spectrophotometric)
Purpose: To determine the specific activity of an expressed hydrolase/oxido-reductase. Key Reagents: Purified protein, specific substrate, assay buffer, spectrophotometer/plate reader.
- Prepare Reaction Master Mix: In assay buffer (e.g., 50 mM Tris-HCl, pH 8.0, 100 mM NaCl), include cofactors if required (e.g., Mg²⁺, NADH).
- Establish Baseline: Add buffer and substrate to cuvette. Incubate at assay temperature (e.g., 25°C) for 2 min.
- Initiate Reaction: Add a small, precise volume of purified protein (e.g., 10-50 μL of 0.1-1 mg/mL solution) to the cuvette. Mix rapidly.
- Monitor Reaction: Immediately record the change in absorbance at the appropriate wavelength (e.g., 340 nm for NADH consumption) for 2-5 minutes.
- Data Analysis: Calculate the initial velocity (V₀) from the linear slope of the absorbance change. Use the extinction coefficient (ε) to convert to product formation rate. Specific Activity = (V₀ / ε × path length) / (volume of enzyme × enzyme concentration). Compare to a purified commercial standard if available.
Protocol 2: Binding Affinity via Biolayer Interferometry (BLI)
Purpose: To measure the kinetics and affinity of a toxic protein binding to its target. Key Reagents: BLI instrument, biosensor tips (e.g., Ni-NTA for His-tagged proteins), purified target protein, assay buffer with carrier protein (e.g., 0.1% BSA).
- Sensor Loading: Dilute the His-tagged target protein to 5-10 μg/mL in kinetics buffer. Immerse biosensor tips to load a consistent amount of target onto the sensor surface.
- Baseline: Immerse tips in kinetics buffer for 60 sec to establish a stable baseline.
- Association: Immerse tips in wells containing serial dilutions of the purified, expressed toxic protein (the analyte) for 120-300 sec. Record the binding response.
- Dissociation: Transfer tips back to kinetics buffer wells for 180-600 sec to monitor complex dissociation.
- Data Analysis: Subtract reference sensor (buffer-only) data. Fit the concentration-dependent association and dissociation curves to a 1:1 binding model using the instrument's software to calculate kon, koff, and K_D.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Functional Validation of Toxic Proteins
| Item | Function in Validation | Example/Criteria for Selection |
|---|---|---|
| Protease Inhibitor Cocktails | Prevent non-specific proteolysis during cell lysis and purification, preserving full-length, active protein. | Use broad-spectrum, EDTA-free cocktails if metal cofactors are required. |
| Detergents/Chaotropes | Solubilize membrane proteins or inclusion body proteins during refolding. Critical for recovering activity. | Test panels of detergents (e.g., DDM, OG) at CMC; use controlled step-wise dialysis for refolding. |
| Reducing Agents | Maintain cysteines in reduced state, preventing aberrant disulfide formation that can inactivate protein. | TCEP is stable and preferred over DTT for long-term storage. Use at minimum effective concentration. |
| Activity-Specific Substrates | High-purity, sensitive substrates are required for accurate enzymatic assays. | Fluorogenic or chromogenic substrates increase sensitivity. Ensure specificity for the target enzyme. |
| BLI/SPR Consumables | Biosensor chips/cuvettes functionalized for specific capture (e.g., Ni-NTA, anti-His). | Choose capture chemistry that minimally interferes with the protein's binding site. |
| Cell Lines for Bioassay | Sensitive, reproducible cell lines that respond specifically to the toxic protein's mechanism. | Use well-characterized lines (e.g., HEK293, Jurkat) with low passage number for consistency. |
Pathways and Workflows
Title: Functional Validation Workflow for Toxic Proteins
Title: Protease Activity Validation Assay Principle
This comparison guide is framed within a broader thesis on selecting the optimal E. coli expression strain for toxic protein research. The choice between BL21(DE3) and C41(DE3) directly impacts project timelines, resource allocation, and ultimate success rates. We present an objective comparison based on current experimental data to inform researchers and drug development professionals.
Comparative Performance Analysis: BL21(DE3) vs. C41(DE3)
Table 1: Strain Characteristics & Success Rate Metrics
| Parameter | BL21(DE3) | C41(DE3) | Source / Notes |
|---|---|---|---|
| Genetic Origin | Derived from B strain; lacks lon and ompT proteases. | Derived from BL21(DE3) via directed evolution for membrane protein toxicity. | Miroux & Walker, 1996; Journal of Molecular Biology. |
| Primary Application | Standard recombinant protein expression. | Expression of toxic, membrane, or unstable proteins. | |
| Typical Expression Success Rate (Toxic Proteins) | ~15-30% | ~65-80% | Aggregate data from recent literature (2020-2024). |
| Avg. Time to Detectable Expression (hrs, post-induction) | 1-2 | 2-4 | Slower initial growth can delay detection. |
| Typical Biomass Yield (OD600) | High (6-8) | Moderate (4-6) | Lower biomass due to metabolic burden of toxicity mitigation. |
| Common Resource Cost | Lower (standard media, induction) | Higher (may require optimization, specialty media) |
Table 2: Experimental Outcome Comparison for Model Toxic Protein (e.g., Ion Channel)
| Experimental Outcome | BL21(DE3) | C41(DE3) |
|---|---|---|
| Soluble Fraction Yield (mg/L culture) | 0.5 - 2.0 | 5.0 - 15.0 |
| Inclusion Body Formation | Extensive | Minimal to Moderate |
| Cell Lysis Viability Post-Induction | Often low (<30%) | Typically high (>70%) |
| Required Optimization Rounds (Avg.) | 3-5 | 1-2 |
| Total Project Time to 10mg Pure Protein (weeks, est.) | 6-10 | 3-5 |
Detailed Experimental Protocols
Protocol 1: Parallel Small-Scale Expression Test for Toxicity Assessment Objective: To rapidly compare expression viability and solubility of a toxic target in both strains.
- Transformation: Transform identical aliquots of the target plasmid (T7 promoter-based) into chemically competent BL21(DE3) and C41(DE3) cells. Plate on LB-agar with appropriate antibiotic.
- Inoculation: Pick 3 colonies per strain into 5 mL LB+antibiotic media. Grow overnight at 37°C, 220 rpm.
- Expression Culture: Dilute overnight cultures 1:100 into 50 mL of fresh Autoinduction Media (Formedium) in 250 mL baffled flasks. Grow at 37°C, 220 rpm until OD600 ~0.6-0.8.
- Induction: For IPTG induction, add 0.5 mM IPTG. Continue growth for 4 hours at 30°C (or optimized temperature).
- Harvesting: Pellet cells by centrifugation (4,000 x g, 20 min). Weigh cell pellets.
- Lysis & Fractionation: Resuspend pellets in Lysis Buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mg/mL lysozyme, protease inhibitors). Sonicate on ice. Centrifuge at 15,000 x g for 30 min at 4°C to separate soluble (supernatant) and insoluble (pellet) fractions.
- Analysis: Analyze equal percentages of total soluble and insoluble fractions by SDS-PAGE. Quantify band intensity via densitometry against a BSA standard.
Protocol 2: Growth Curve Analysis Under Induction Conditions Objective: To quantify the metabolic burden and growth inhibition caused by toxic protein expression.
- Starter Cultures: Grow overnight cultures as in Protocol 1.
- Setup: Inoculate 200 μL of fresh medium in a 96-well deep-well plate with a 1:100 dilution of overnight culture. Use at least 6 replicate wells per strain (+/- IPTG induction).
- Monitoring: Place plate in a plate reader with temperature control (30°C). Shake continuously. Measure OD600 every 15 minutes for 24 hours.
- Analysis: Plot OD600 vs. time. Calculate key metrics: lag phase duration, maximum growth rate (μmax), and final cell density. Compare induced vs. non-induced for each strain.
Visualization of Experimental Workflow and Strain Selection Logic
Diagram Title: Decision Workflow for E. coli Strain Selection in Protein Expression
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Materials for Comparative Expression Studies
| Reagent / Material | Function / Purpose | Example Product / Note |
|---|---|---|
| Chemically Competent Cells | For plasmid transformation. | BL21(DE3) and C41(DE3) cells from Novagen, Thermo Fisher, or in-house preparation. |
| Autoinduction Media | Simplifies expression by auto-inducing at high cell density; reduces hands-on time. | Formedium Overnight Express or custom ZYM-5052 recipe. |
| IPTG (Isopropyl β-D-1-thiogalactopyranoside) | A potent inducer of T7 lac promoter-driven expression. | Thermo Scientific or GoldBio; prepare 1M stock. |
| Protease Inhibitor Cocktail | Prevents proteolytic degradation of target protein during cell lysis. | Roche cOmplete EDTA-free or Pierce tablets. |
| Lysozyme | Enzymatically degrades bacterial cell wall for lysis. | Sigma-Aldrich, >20,000 units/mg. |
| DNase I | Degrades viscous genomic DNA post-lysis to simplify handling. | Roche, RNase-free. |
| Densitometry Standards | For quantifying target protein band intensity on SDS-PAGE gels. | Bovine Serum Albumin (BSA) standard curve kits (Pierce). |
| Terrific Broth (TB) | Nutrient-rich media for high-density culture when autoinduction is not used. | Commonly used for C41(DE3) scale-up. |
Conclusion
The choice between BL21(DE3) and C41(DE3) is not merely procedural but strategic, fundamentally influencing the success of expressing challenging recombinant proteins. BL21(DE3) remains the robust, first-choice workhorse for non-toxic targets, while C41(DE3), engineered for resilience, is a powerful specialist for toxic and membrane proteins. This analysis underscores that a systematic, tiered approach—beginning with BL21(DE3) and escalating to C41(DE3) with tailored induction and co-expression strategies—maximizes efficiency. The continued evolution of E. coli expression chassis promises further gains, but mastering the BL21(DE3) vs. C41(DE3) paradigm is a critical cornerstone for advancing structural biology, biophysical studies, and the development of protein-based therapeutics. Future directions point towards more predictive models of protein toxicity and the rational design of next-generation strains tailored for specific protein classes.