Unfolding the ATF6-GRP78 Chaperone System: A Comprehensive Guide to Endoplasmic Reticulum Protein Folding and Stress Signaling
This article provides a detailed exploration of the ATF6-GRP78 chaperone system, a central regulator of endoplasmic reticulum (ER) proteostasis and the unfolded protein response (UPR).
Unfolding the ATF6-GRP78 Chaperone System: A Comprehensive Guide to Endoplasmic Reticulum Protein Folding and Stress Signaling
Abstract
This article provides a detailed exploration of the ATF6-GRP78 chaperone system, a central regulator of endoplasmic reticulum (ER) proteostasis and the unfolded protein response (UPR). Aimed at researchers and drug development professionals, it covers the foundational molecular biology of ATF6 activation and GRP78 function, current methodologies for studying this system, common experimental challenges and optimization strategies, and validation techniques for assessing its role in disease models. The content synthesizes the latest research to highlight this pathway's critical implications for neurodegenerative diseases, cancer, and metabolic disorders, offering a roadmap for therapeutic targeting.
The ATF6-GRP78 Axis: Core Mechanisms of ER Stress Sensing and Protein Folding
Within the complex machinery of the endoplasmic reticulum (ER), the maintenance of proteostasis is paramount. The dynamic interplay between the ER-resident chaperone GRP78/BiP and the stress-sensor transcription factor ATF6 forms a critical regulatory node of the unfolded protein response (UPR). This whitepaper defines the molecular architecture and constitutive functions of these two core players, framing them within the broader context of the ATF6-GRP78 chaperone system in protein folding research. Understanding this dyad is fundamental for developing therapeutic interventions in diseases characterized by ER stress, including neurodegeneration, cancer, and metabolic disorders.
Molecular Structure and Domains
ATF6 (Activating Transcription Factor 6)
ATF6 is a type II transmembrane protein localized to the ER. It functions as a potent ER stress sensor and transducer. Its structure is modular, with distinct domains governing its localization, regulation, and transcriptional activity.
Table 1: Structural Domains of ATF6 (ATF6α Isoform)
| Domain | Amino Acid Region (Approx.) | Structural/Functional Description |
|---|---|---|
| Luminal Domain (N-terminus) | 1-373 | Senses ER stress; contains BiP-binding sites. Undergoes conformational change upon BiP dissociation. |
| Transmembrane Domain | 374-397 | Anchors ATF6 in the ER membrane. |
| Cytosolic Domain | 398-670 | Contains basic-leucine zipper (bZIP) motif for DNA binding and dimerization. Cleaved by S1P and S2P proteases to release the active cytosolic fragment (ATF6f). |
| bZIP Domain | ~520-590 | Mediates DNA binding to ERSE (ER Stress Response Element) and dimerization with other bZIP proteins (e.g., XBP1, NF-Y). |
| Golgi Localization Signal (GLS) | Within Luminal Domain | Exposed upon BiP dissociation, targets ATF6 to the Golgi for proteolytic activation. |
GRP78/BiP (Glucose-Regulated Protein 78 kDa / Immunoglobulin Heavy Chain-Binding Protein)
GRP78 (HSPA5) is a central ER-resident chaperone of the HSP70 family. It is a master regulator of ER homeostasis, integrating folding, quality control, and signaling functions.
Table 2: Structural Domains of GRP78/BiP
| Domain | Amino Acid Region | Structural/Functional Description |
|---|---|---|
| Nucleotide-Binding Domain (NBD) | ~1-386 | Binds and hydrolyzes ATP. ATP/ADP cycling governs substrate binding affinity. Contains key residues for ATPase activity (e.g., T229). |
| Substrate-Binding Domain (SBD) | ~387-654 | Binds hydrophobic peptide segments of unfolded/misfolded client proteins. Comprises a β-sandwich subdomain (SBDβ) for peptide binding and an α-helical lid (SBDα). |
| Linker Region | Connects NBD & SBD | Transduces conformational changes between domains during the allosteric cycle. |
| ER Retention Signal | C-terminal KDEL | Tethers BiP to the ER lumen; retrieved from the Golgi via KDEL receptors. |
Basal Functions and Regulatory Interplay
GRP78/BiP: The Central Chaperone
Under homeostatic conditions, GRP78/BiP executes essential functions:
- De Novo Protein Folding: Binds nascent polypeptide chains entering the ER, preventing aggregation and facilitating folding.
- Protein Translocation: Acts as a seal and ratchet at the translocon pore, assisting in polypeptide import.
- Calcium Buffering: Exhibits low-affinity, high-capacity calcium binding, contributing to ER luminal calcium stores.
- Master UPR Repression: Directly binds and inactivates the luminal domains of all three ER stress sensors: ATF6, IRE1α, and PERK.
ATF6: The Stress-Responsive Transcription Factor
Under non-stress conditions, ATF6 is held inactive in the ER membrane via direct binding of GRP78 to its luminal domain. Its basal activity is minimal.
The Activation Cycle: ATF6 and GRP78 Dissociation
Upon ER stress (e.g., accumulation of unfolded proteins), GRP78 is competitively sequestered by unfolded clients, leading to its dissociation from ATF6. This exposes the Golgi Localization Signal (GLS) on ATF6.
Diagram 1: ATF6 Activation by GRP78 Sequestration
The liberated ATF6 traffics to the Golgi apparatus, where it is sequentially cleaved by Site-1 Protease (S1P) and Site-2 Protease (S2P). This regulated intramembrane proteolysis releases the soluble, active N-terminal cytosolic fragment (ATF6f), which translocates to the nucleus.
Core Experimental Protocols
Protocol 1: Monitoring ATF6 Activation via Immunoblotting Objective: Detect the proteolytic cleavage of full-length ATF6 (~90 kDa) to its active cytosolic fragment ATF6f (~50 kDa).
- Cell Treatment & Lysis: Treat cells (e.g., HEK293, HeLa) with ER stress inducer (e.g., 2-5 µg/mL Tunicamycin, 1-10 µM Thapsigargin) for 1-8 hours. Harvest cells in RIPA buffer with protease inhibitors.
- Subcellular Fractionation (Optional but Recommended):
- Use a commercial cytoplasmic/nuclear extraction kit.
- Confirm purity using markers (e.g., Lamin B1 for nucleus, α-Tubulin for cytoplasm).
- Immunoblotting:
- Load 20-40 µg of total protein or nuclear extract per lane on a 4-12% Bis-Tris gel.
- Transfer to PVDF membrane.
- Block with 5% non-fat milk in TBST.
- Probe with primary antibodies:
- Anti-ATF6α (Full-length): Rabbit monoclonal (clone EPR4211), 1:1000, incubation at 4°C overnight.
- Anti-ATF6α (Cleaved/Active): Mouse monoclonal (clone 4F2), 1:500, detects ATF6f.
- Use appropriate HRP-conjugated secondary antibodies (1:5000).
- Develop with chemiluminescent substrate and image.
Protocol 2: Measuring GRP78-Binding Dynamics by Co-Immunoprecipitation (Co-IP) Objective: Assess the physical interaction between GRP78 and ATF6 under stress vs. non-stress conditions.
- Cell Treatment & Lysis: Treat cells as in Protocol 1. Lyse in non-denaturing IP lysis buffer (e.g., 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, with protease inhibitors). Avoid SDS or strong detergents.
- Pre-clearing: Incubate lysate with 20 µL of Protein A/G agarose beads for 30 min at 4°C. Pellet beads, keep supernatant.
- Immunoprecipitation:
- Incubate 500 µg of pre-cleared lysate with 2 µg of anti-GRP78 antibody (e.g., clone C50B12) or species-matched IgG control for 2 hours at 4°C.
- Add 40 µL of Protein A/G beads and incubate overnight at 4°C with gentle rotation.
- Washing and Elution: Pellet beads, wash 3x with ice-cold lysis buffer. Elute bound proteins in 2X Laemmli sample buffer by heating at 95°C for 5 min.
- Immunoblot Analysis: Run eluate on SDS-PAGE. Probe blot sequentially for ATF6 (to detect co-precipitated protein) and GRP78 (to confirm IP efficiency).
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Studying the ATF6-GRP78 System
| Reagent / Material | Function / Application | Example (Supplier) |
|---|---|---|
| ER Stress Inducers | Experimentally induce ER stress to activate the UPR pathways. | Tunicamycin (N-glycosylation inhibitor; Sigma T7765), Thapsigargin (SERCA pump inhibitor; Sigma T9033), DTT (reducing agent; disrupts disulfides). |
| ATF6 Activation Inhibitor | Specifically blocks S1P/S2P-mediated cleavage of ATF6 in the Golgi. | AEBSF (4-(2-Aminoethyl)benzenesulfonyl fluoride hydrochloride; Sigma A8456). |
| Anti-ATF6α Antibodies | Detect full-length and cleaved ATF6 via WB, IF, IP. | Full-length: Abcam ab122897 (EPR4211); Cleaved/Active: Sigma A7826 (4F2). |
| Anti-GRP78/BiP Antibodies | Detect GRP78 expression (stress marker) and for Co-IP. | Cell Signaling Technology #3177 (C50B12) for WB/IP; Abcam ab21685 for IF. |
| GRP78/BiP siRNA/shRNA | Knockdown GRP78 to study its essential role in ATF6 regulation. | SMARTpool siRNA (Dharmacon, L-008194-00). |
| ATF6 Reporter Plasmid | Measure ATF6 transcriptional activity luciferase-based. | p5xATF6-GL3 (Addgene, plasmid 11976). |
| Subcellular Fractionation Kit | Isolate nuclear extracts to analyze ATF6f translocation. | NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher, 78833). |
| Protease Inhibitor Cocktail | Prevent degradation of ATF6 and other proteins during lysis. | cOmplete, EDTA-free (Roche, 4693132001). |
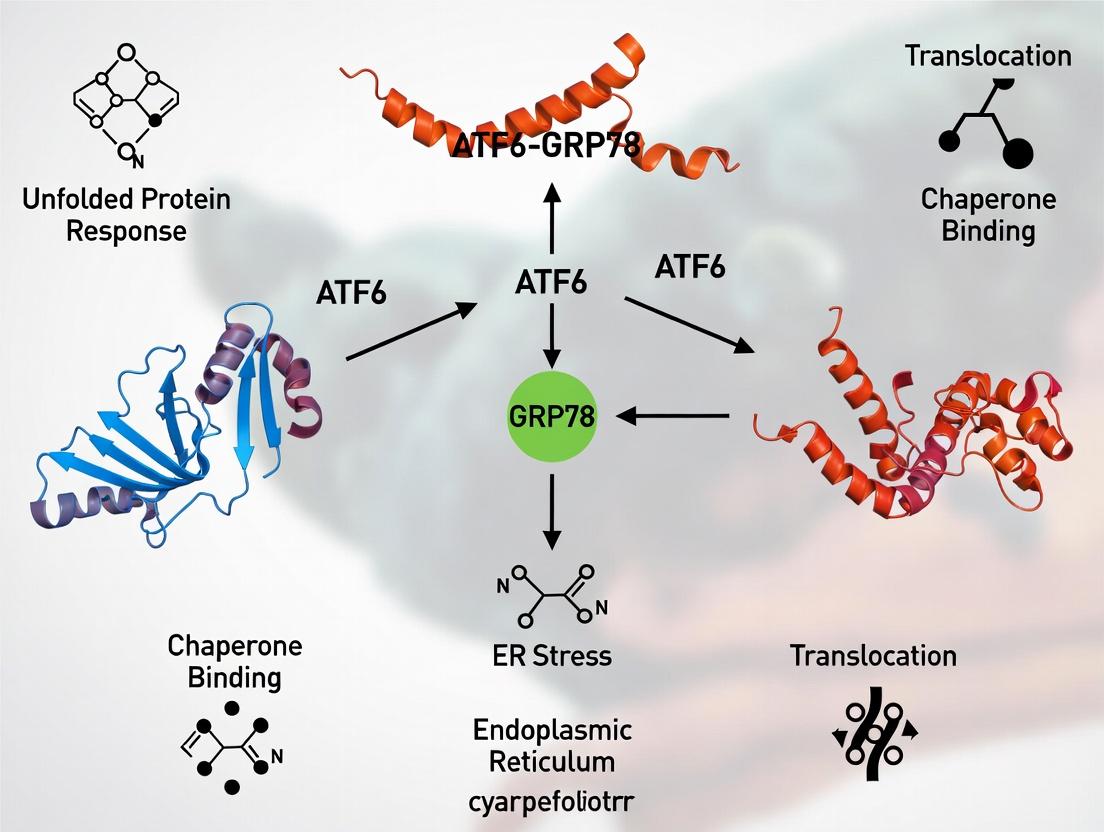
Within the endoplasmic reticulum (ER), the fidelity of protein folding is monitored by a network of chaperones and sensors. Central to this network is the chaperone GRP78 (BiP/HSPA5) and its dynamic interaction with the transmembrane sensor ATF6 (Activating Transcription Factor 6). This "dance" in the ER lumen—the cyclic binding and release—serves as the fundamental regulator of ATF6 inactivation under basal conditions and its release upon ER stress. This whitepaper, framed within the broader thesis of chaperone-mediated folding regulation, details the molecular mechanics, quantitative parameters, and experimental interrogation of this critical system for researchers and drug development professionals.
Molecular Mechanism: The Binding-Release Cycle
ATF6 exists as a type II transmembrane protein. Its ER-luminal domain is constitutively bound by GRP78 under non-stress conditions. This interaction physically sequesters ATF6, preventing its dimerization and trafficking to the Golgi apparatus.
Key Mechanistic Steps:
- Basal State (Inactivation): GRP78 binds to the luminal domain of ATF6 with high affinity, tethering it in the ER.
- ER Stress Induction: Accumulation of unfolded proteins competes for GRP78 binding, drawing chaperones away from ATF6.
- Release and Activation: The dissociation of GRP78 unmasks intra- and intermolecular interactions, leading to ATF6 dimerization, packaging into COPII vesicles, and transport to the Golgi.
- Proteolytic Cleavage: In the Golgi, ATF6 is processed by Site-1 and Site-2 Proteases (S1P, S2P), releasing its cytosolic N-terminal domain.
- Transcriptional Reprogramming: The released fragment translocates to the nucleus, acting as a transcription factor to upregulate genes encoding ER chaperones (including GRP78), foldases, and components of ER-associated degradation (ERAD).
Diagram 1: GRP78-ATF6 Regulatory Cycle Pathway
Quantitative Binding & Kinetic Data
Table 1: Key Quantitative Parameters of the ATF6-GRP78 Interaction
| Parameter | Value / Range | Experimental Method | Significance |
|---|---|---|---|
| GRP78-ATF6 Kd (Basal) | ~ 0.5 - 2.0 µM | Surface Plasmon Resonance (SPR), Isothermal Titration Calorimetry (ITC) | Defines high-affinity interaction under non-stress conditions. |
| GRP78-Unfolded Protein Kd | ~ 0.1 - 1.0 µM (varies by substrate) | Fluorescence Anisotropy, ITC | Higher affinity for unfolded clients drives competitive dissociation from ATF6. |
| ATF6 Golgi Transit Time Post-Stress | 30 - 90 minutes | Cycloheximide chase, Immunofluorescence time-course | Kinetics of activation after stress induction. |
| S1P/S2P Cleavage Half-time | ~ 15 - 30 minutes (post-Golgi arrival) | Western blot analysis of cleavage intermediates | Rate-limiting step for nuclear fragment generation. |
| GRP78 mRNA Induction Fold-Change | 3x - 10x (cell-type dependent) | qPCR, RNA-Seq | Measures transcriptional output of activated ATF6 pathway. |
Key Experimental Protocols
Co-Immunoprecipitation (Co-IP) to Assess GRP78-ATF6 Association
Objective: To validate the physical interaction between GRP78 and ATF6 under basal and ER stress conditions.
- Cell Lysis: Lyse HEK293 or HeLa cells in mild non-denaturing lysis buffer (e.g., 1% digitonin or CHAPS in TBS) supplemented with protease inhibitors.
- Pre-Clearance: Incubate lysate with control IgG and Protein A/G beads for 1h at 4°C. Pellet beads, retain supernatant.
- Immunoprecipitation: Incubate supernatant with anti-ATF6 or anti-GRP78 antibody (or species-matched control IgG) overnight at 4°C with gentle rotation.
- Bead Capture: Add Protein A/G agarose beads for 2h. Pellet and wash beads 4x with lysis buffer.
- Elution & Analysis: Elute proteins in 2X Laemmli buffer at 95°C for 5 min. Analyze by SDS-PAGE and Western blot, probing for the co-precipitated partner (GRP78 or ATF6).
Monitoring ATF6 Trafficking via Immunofluorescence
Objective: To visualize the stress-induced translocation of ATF6 from the ER to the Golgi.
- Cell Culture & Stress: Seed cells on glass coverslips. Treat with ER stressor (e.g., 2µg/mL Tunicamycin, 5mM DTT) or vehicle for a predetermined time (e.g., 90 min).
- Fixation & Permeabilization: Fix with 4% paraformaldehyde for 15 min, permeabilize with 0.1% Triton X-100 for 10 min.
- Staining: Block with 5% BSA. Incubate with primary antibodies: anti-ATF6 (luminal domain) and a Golgi marker (e.g., anti-GM130). Follow with appropriate fluorescent secondary antibodies (e.g., Alexa Fluor 488, 594).
- Imaging & Analysis: Image using confocal microscopy. Co-localization analysis (e.g., Pearson's coefficient) between ATF6 and the Golgi marker quantifies trafficking.
Reporter Gene Assay for ATF6 Transcriptional Activity
Objective: To functionally measure the activation of the ATF6 pathway.
- Reporter Construct: Transfect cells with a plasmid containing multiple ER stress response elements (ERSE) upstream of a luciferase gene (e.g., Firefly luciferase).
- Normalization: Co-transfect with a constitutive promoter-driven Renilla luciferase plasmid for normalization.
- Induction & Lysis: Induce ER stress for 8-16 hours. Lyse cells using a dual-luciferase assay buffer.
- Measurement: Measure Firefly and Renilla luciferase signals sequentially using a luminometer. The Firefly/Renilla ratio reflects ATF6 pathway activity.
Logical Relationship of Key Findings
Diagram 2: Logic of ATF6 Activation by Competitive Displacement
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Investigating the ATF6-GRP78 System
| Reagent / Material | Function / Application | Key Consideration |
|---|---|---|
| Tunicamycin | N-linked glycosylation inhibitor. Classic pharmacological inducer of ER stress and ATF6 activation. | Use low concentrations (0.5-5 µg/mL); highly cytotoxic over long periods. |
| Dithiothreitol (DTT) | Reducing agent. Causes ER stress by disrupting disulfide bond formation, rapidly inducing ATF6 processing. | Typical dose 1-5mM. Effects are rapid but can be pleiotropic. |
| ATF6α (D4G8) Rabbit mAb | Detects full-length (p90) and cleaved nuclear (p50) ATF6 by Western blot. Common for tracking activation. | Optimal for human, mouse, rat samples. Does not cross-react with ATF6β. |
| GRP78/BiP (C50B12) Rabbit mAb | Standard antibody for detecting GRP78 in Western blot, IP, and IF. Monitors chaperone induction. | Confirm expected molecular weight (~78 kDa) to distinguish from other HSP70s. |
| ER-Tracker Dyes | Live-cell staining of the endoplasmic reticulum. Used in imaging studies to localize ATF6 pre-activation. | Compatible with fixation for some protocols; check specifications. |
| Site-1 Protease (MBTPS1) Inhibitor (PF-429242) | Specific, cell-active inhibitor of S1P. Blocks the final activation step of ATF6, used to confirm processing. | Used at ~10-50 µM. Accumulates uncleaved, Golgi-localized ATF6. |
| ATF6 Reporter Plasmid (p5xATF6-GL3) | Luciferase construct driven by ATF6-responsive elements. Gold standard for functional pathway readout. | Requires co-transfection with a Renilla control for normalization. |
| Recombinant GRP78 Protein | For in vitro binding assays (SPR, ITC) to determine binding constants with ATF6 luminal domain peptides. | Ensure protein is purified, refolded, and ATPase activity is characterized. |
This whitepaper provides an in-depth technical guide to the signal transduction pathway culminating in the proteolytic activation of the transcription factor ATF6. The content is framed within the broader thesis of the ATF6-GRP78 chaperone system as a central regulatory node in cellular protein folding homeostasis. In the endoplasmic reticulum (ER), the accumulation of unfolded or misfolded proteins disrupts this homeostasis, triggering the Unfolded Protein Response (UPR). ATF6 is a key UPR sensor, and its activation represents a critical step in restoring ER function. This document details the molecular journey of ATF6 from an ER-transmembrane protein to a liberated, transcriptionally active nuclear factor, with a focus on quantitative insights and experimental approaches for researchers and drug development professionals.
The Core Signaling Pathway: A Stepwise Mechanism
The activation of ATF6 is a tightly regulated, multi-compartment process.
2.1. ER Stress Sensing and Mobilization Under non-stress conditions, ATF6 (approx. 90 kDa type II transmembrane protein) is retained in the ER lumen through binding to the chaperone GRP78 (BiP). The dissociation constant (Kd) for the ATF6-GRP78 interaction is estimated to be in the low nanomolar range, ensuring stable complex formation. Upon ER stress, GRP78 is sequestered by a rising load of misfolded proteins, leading to its release from ATF6.
2.2. ER-to-Golgi Trafficking GRP78 dissociation unmasks ER export motifs on ATF6, facilitating its packaging into COPII-coated vesicles. Quantitative live-cell imaging indicates that ATF6 translocates to the Golgi apparatus with a half-time (t1/2) of approximately 15-30 minutes post-stress induction.
2.3. Proteolytic Activation in the Golgi Within the Golgi, ATF6 encounters two resident proteases:
- Site-1 Protease (S1P): A serine protease that cleaves ATF6 in its luminal domain.
- Site-2 Protease (S2P): A metalloprotease that performs intramembrane cleavage.
This regulated intramembrane proteolysis (RIP) releases the soluble N-terminal cytoplasmic domain of ATF6 (ATF6f, ~50 kDa), which contains a basic leucine zipper (bZIP) DNA-binding domain.
2.4. Nuclear Translocation and Transcriptional Regulation ATF6f translocates to the nucleus (driven by a nuclear localization signal) and forms homodimers or heterodimers with other bZIP proteins. It binds to ER Stress Response Elements (ERSE, consensus: CCAAT-N9-CCACG) and UPRE promoters, upregulating genes encoding ER chaperones (e.g., GRP78, GRP94), foldases, and components of ER-associated degradation (ERAD).
Table 1: Key Quantitative Parameters in ATF6 Activation
| Parameter | Approximate Value / Range | Measurement Method | Biological Context |
|---|---|---|---|
| ATF6-GRP78 Kd | 1-10 nM | Surface Plasmon Resonance (SPR), Co-IP | ER homeostasis |
| Golgi Trafficking t1/2 | 15-30 min | Fluorescence Recovery After Photobleaching (FRAP) | Post-stress mobilization |
| S1P Cleavage Site | Arginine-X-Arginine (RXR) motif | Mutagenesis & Mass Spectrometry | Golgi compartment |
| ATF6f Size | ~50 kDa | Western Blot (SDS-PAGE) | Cleavage product |
| ERSE Binding Affinity (Kd) | 10-20 nM | Electrophoretic Mobility Shift Assay (EMSA) | Transcriptional activation |
Diagram 1: ATF6 Activation Pathway from ER to Nucleus (89 characters)
Detailed Experimental Protocols
3.1. Monitoring ATF6 Trafficking via Immunofluorescence & Confocal Microscopy
- Objective: Visualize subcellular localization of ATF6 pre- and post-ER stress.
- Procedure:
- Seed cells (e.g., HeLa, HEK293) on glass coverslips.
- Induce ER stress using 2-5 µM thapsigargin (SERCA inhibitor) or 1-5 µg/mL tunicamycin (N-glycosylation inhibitor) for 1-3 hours.
- Fix cells with 4% paraformaldehyde (PFA) for 15 min, permeabilize with 0.1% Triton X-100.
- Block with 5% BSA in PBS for 1 hour.
- Incubate with primary antibodies (mouse anti-ATF6, rabbit anti-GM130 [Golgi marker]) diluted in blocking buffer overnight at 4°C.
- Wash and incubate with fluorescent secondary antibodies (e.g., Alexa Fluor 488 anti-mouse, Alexa Fluor 555 anti-rabbit) for 1 hour at RT.
- Stain nuclei with DAPI, mount, and image using a confocal microscope.
- Key Analysis: Quantify co-localization of ATF6 signal with Golgi marker using Pearson's correlation coefficient.
3.2. Detecting ATF6 Cleavage via Western Blot Analysis
- Objective: Assess ATF6 proteolytic processing by detecting full-length (p90ATF6) and cleaved (p50ATF6f) forms.
- Procedure:
- Treat cells with ER stress inducers (as above) for varying time points (0, 0.5, 1, 2, 4 h).
- Lyse cells in RIPA buffer containing protease inhibitors.
- Resolve 20-30 µg of total protein on a 10% SDS-PAGE gel.
- Transfer to PVDF membrane.
- Block with 5% non-fat milk in TBST.
- Probe with anti-ATF6 N-terminal antibody (to detect both full-length and cleaved form) at 4°C overnight.
- Incubate with HRP-conjugated secondary antibody, develop with ECL reagent, and visualize.
- Key Analysis: The appearance of the ~50 kDa ATF6f band indicates successful activation.
3.3. Assessing Transcriptional Activity via Luciferase Reporter Assay
- Objective: Quantify ATF6-dependent transcriptional activation.
- Procedure:
- Co-transfect cells with a plasmid containing a firefly luciferase gene under an ERSE promoter and a Renilla luciferase control plasmid for normalization.
- After 24 hours, induce ER stress.
- At 12-24 hours post-induction, lyse cells and measure firefly and Renilla luciferase activities using a dual-luciferase assay kit.
- Calculate the ratio of firefly/Renilla luminescence.
- Key Analysis: Fold increase in normalized luciferase activity relative to untreated controls indicates ATF6 transcriptional output.
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for ATF6 Pathway Research
| Reagent/Category | Example Product/Specifics | Primary Function in ATF6 Research |
|---|---|---|
| ER Stress Inducers | Thapsigargin, Tunicamycin, Dithiothreitol (DTT) | Induce ER protein folding burden, triggering GRP78 release and ATF6 mobilization. |
| Protease Inhibitors | AEBSF (S1P inhibitor), 1,10-Phenanthroline (Metalloprotease/S2P inhibitor) | Chemically validate the requirement for S1P/S2P in ATF6 cleavage. |
| Antibodies (Anti-ATF6) | Monoclonal (e.g., Clone 1G7), Polyclonal (N-terminal vs. C-terminal specific) | Detect full-length and cleaved ATF6 via WB, IF, or IP. N-terminal antibodies recognize both forms. |
| Antibodies (Markers) | Anti-GRP78/BiP, Anti-GM130, Anti-Calnexin, Anti-KDEL | Mark ER/Golgi compartments for co-localization studies and monitor GRP78 expression. |
| Luciferase Reporter | pGL4-ERSE-Luciferase, pRL-TK (Renilla control) | Quantify ATF6-driven transcriptional activity in live cells. |
| Expression Vectors | Plasmid encoding FLAG/GFP-tagged ATF6 (wild-type & mutant) | Overexpress or mutate ATF6 to study trafficking, cleavage, and function. |
| siRNA/shRNA | Targeted against ATF6, S1P, S2P, GRP78 | Knock down specific pathway components to establish genetic necessity. |
| Chemical Chaperones | 4-Phenylbutyric Acid (4-PBA), Tauroursodeoxycholic Acid (TUDCA) | Ameliorate ER stress; used as negative controls or therapeutic probes. |
Diagram 2: Experimental Workflow for ATF6 Pathway Analysis (78 characters)
The precise signal transduction from ER stress to ATF6 activation is a paradigm of organelle-specific proteolytic signaling. Within the thesis of the ATF6-GRP78 system as guardians of proteostasis, this pathway represents a dynamic feedback loop: stress reduces folding capacity, GRP78 is diverted, ATF6 activates, and its target genes (including GRP78 itself) work to restore folding capacity. This makes the pathway a compelling target for drug discovery. Modulators that can potentiate ATF6 activation (e.g., in degenerative diseases involving chronic proteotoxicity) or temporarily inhibit it (e.g., in cancers exploiting the UPR for survival) represent promising therapeutic avenues. Continued quantitative dissection of its trafficking kinetics, protease specificity, and transcriptional network is essential for translating this knowledge into targeted interventions.
Within the broader thesis on the ATF6-GRP78 chaperone system in protein folding research, this whitepaper elucidates the central role of Activating Transcription Factor 6 (ATF6) as a master regulator of the unfolded protein response (UPR). Upon endoplasmic reticulum (ER) stress, ATF6 translocates to the Golgi, is cleaved, and its cytosolic fragment (ATF6f) translocates to the nucleus to orchestrate the transcription of a network of genes dedicated to restoring ER proteostasis. This guide provides an in-depth technical analysis of key ATF6 target genes—notably GRP78 (BiP), XBP1, and molecular chaperones—and their integrated function in mitigating ER stress.
The ATF6 Signaling Pathway: From Activation to Transcription
Pathway Diagram
Title: ATF6 Activation and Transcriptional Regulation Pathway
Key Regulatory Steps
- Homeostatic State: ATF6 (p90) is retained in the ER membrane, bound by the chaperone GRP78/BiP.
- ER Stress Induction: Accumulation of unfolded proteins sequesters GRP78, releasing ATF6.
- Vesicular Transport: ATF6 is packaged into COPII vesicles and transported to the Golgi apparatus.
- Regulated Intramembrane Proteolysis (RIP): In the Golgi, ATF6 is cleaved sequentially by Site-1 Protease (S1P) and Site-2 Protease (S2P), releasing its cytosolic fragment (ATF6f, p50).
- Nuclear Translocation & DNA Binding: ATF6f translocates to the nucleus and binds to ER Stress Response Elements (ERSE: CCAAT-N9-CCACG) in the promoters of target genes.
- Transcriptional Activation: ATF6f, often in complex with general transcription factors (e.g., NF-Y/CBF), drives the expression of UPR target genes.
Core ATF6 Target Genes and Their Functions
ATF6 directly upregulates a suite of genes that collectively enhance the ER's folding, quality control, and clearance capacity.
GRP78 (BiP/HSPA5): The Central ER Chaperone
The HSPA5 gene encoding GRP78 is the canonical ATF6 target. GRP78 is an HSP70-family chaperone that acts as the primary ER luminal sensor for unfolded proteins and a central regulator of all three UPR branches (ATF6, IRE1, PERK).
Functions:
- Polypeptide Binding: Binds hydrophobic patches on nascent/unfolded proteins, preventing aggregation.
- ATPase Activity: Hydrolyzes ATP to drive cycles of client protein binding and release.
- UPR Master Regulator: When bound to unfolded clients, it releases ATF6, IRE1, and PERK, activating the UPR.
XBP1: Amplifying the UPR Signal
ATF6 transcriptionally upregulates XBP1 mRNA. This pre-mRNA is then spliced by the endoribonuclease IRE1α (activated independently by ER stress) to produce the potent transcription factor XBP1s. Thus, ATF6 action amplifies the IRE1/XBP1 arm of the UPR.
Functions of XBP1s:
- Binds to UPRE and ERSE-II promoter elements.
- Drives expression of genes involved in ER-associated degradation (ERAD), lipid biosynthesis, and additional chaperones, expanding the ER's functional capacity.
ER-Resident Chaperones and Foldases
ATF6 upregulates a network of chaperones and enzymes that facilitate protein folding and maturation.
Key Examples:
- GRP94 (HSP90B1): An HSP90-family chaperone specializing in the maturation of client proteins like Toll-like receptors and integrins.
- Protein Disulfide Isomerase (PDI) Family: Catalyzes the formation, breakage, and isomerization of disulfide bonds (e.g., PDI, PDIA6).
- Calnexin & Calreticulin: Lectin chaperones that associate with glycoproteins, promoting folding and quality control.
Functional Integration Diagram
Title: Integrated Network of ATF6 Target Gene Functions
Quantitative Data on ATF6 Target Gene Induction
Table 1: Representative Fold-Induction of Key ATF6 Target Genes Under ER Stress Data compiled from recent studies using thapsigargin (Tg) or tunicamycin (Tm) in mammalian cell lines (e.g., HEK293, HeLa).
| Target Gene | Protein | ER Stressor | Fold Induction (mRNA) | Time to Peak Induction | Primary Assay |
|---|---|---|---|---|---|
| HSPA5 | GRP78/BiP | Tm (2μg/ml) | 8 - 12x | 8 - 16 hrs | qRT-PCR, RNA-seq |
| HSP90B1 | GRP94 | Tg (1μM) | 5 - 8x | 8 - 12 hrs | qRT-PCR |
| XBP1 | XBP1 (unspliced) | Tm (2μg/ml) | 4 - 6x | 4 - 8 hrs | qRT-PCR |
| DNAJB11 | ERdj3/HEDJ | Tg (1μM) | 6 - 10x | 12 hrs | Microarray |
| PDIA4 | Protein Disulfide Isomerase A4 | Tm (2μg/ml) | 3 - 5x | 12 hrs | RNA-seq |
| HERPUD1 | HERP | Tg (1μM) | 10 - 15x | 8 hrs | qRT-PCR |
Table 2: Core ERSE Promoter Elements in Key ATF6 Target Genes
| Target Gene | Canonical ERSE Sequence (5' -> 3') | Position Relative to TSS | Confirmed by |
|---|---|---|---|
| HSPA5 (GRP78) | CCAAT-N9-CCACG | ~70 to -50 | ChIP, Luciferase Reporter |
| HSP90B1 (GRP94) | CCAAT-N9-CCACG | -100 to -80 | EMSA, Mutagenesis |
| XBP1 | CCAAT-N9-CCACG | -120 to -100 | ChIP-seq |
| DNAJB11 | CCAAT-N9-CCACG | -150 to -130 | Reporter Assay |
Experimental Protocols for Investigating ATF6 Function
Protocol: Monitoring ATF6 Cleavage and Nuclear Translocation
Objective: To detect the proteolytic activation and nuclear accumulation of ATF6 in response to ER stress.
Materials: See "The Scientist's Toolkit" (Section 7). Procedure:
- Cell Treatment: Seed HEK293 or HeLa cells in 6-well plates. At 70-80% confluency, treat with 2μM thapsigargin or 2μg/ml tunicamycin in DMSO. Use DMSO alone as vehicle control.
- Fractionation (2-8 hrs post-treatment):
- Harvest cells, wash with PBS.
- Lyse cells in Cytoplasmic Extraction Buffer (CEB: 10 mM HEPES pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.4% NP-40, plus protease inhibitors) on ice for 15 min. Centrifuge at 12,000g for 5 min at 4°C.
- Collect supernatant as Cytoplasmic Fraction.
- Wash pellet with CEB, then resuspend in Nuclear Extraction Buffer (NEB: 20 mM HEPES pH 7.9, 400 mM NaCl, 1 mM EDTA, 10% glycerol, plus inhibitors). Vortex vigorously, incubate on ice for 30 min. Centrifuge at 12,000g for 10 min.
- Collect supernatant as Nuclear Fraction.
- Western Blot Analysis:
- Run 20-40μg of each fraction on 4-12% Bis-Tris gel.
- Transfer to PVDF membrane.
- Probe with primary antibodies: Anti-ATF6α (for full-length p90 and cleaved p50), Anti-Lamin B1 (nuclear marker), Anti-α-Tubulin (cytoplasmic marker).
- Use HRP-conjugated secondary antibodies and chemiluminescent detection. Expected Result: Increased p50 signal in the nuclear fraction post-stress, with corresponding decrease in p90 signal in the cytoplasmic/ER fraction.
Protocol: Chromatin Immunoprecipitation (ChIP) for ATF6 Binding
Objective: To confirm direct binding of ATF6f to the ERSE of a target gene (e.g., HSPA5 promoter).
Procedure:
- Crosslinking & Lysis: Treat cells as in 5.1. After 4 hrs, crosslink with 1% formaldehyde for 10 min at RT. Quench with 125mM glycine. Harvest, wash, and lyse cells in SDS Lysis Buffer.
- Sonication: Sonicate chromatin to shear DNA to fragments of 200-500 bp. Centrifuge to pellet debris.
- Immunoprecipitation: Pre-clear lysate with protein A/G beads. Incubate supernatant overnight at 4°C with Anti-ATF6α antibody or normal IgG control. Add beads, incubate, wash extensively.
- Elution & Reverse Crosslinking: Elute complexes, add NaCl, and heat at 65°C overnight to reverse crosslinks. Treat with Proteinase K.
- DNA Purification & Analysis: Purify DNA using a spin column. Analyze by qPCR with primers flanking the ERSE in the HSPA5 promoter and a control region from a non-target gene. Expected Result: Enrichment of the HSPA5 ERSE amplicon, but not the control region, in the ATF6 ChIP sample from stressed cells.
Research Reagent Solutions: The Scientist's Toolkit
Table 3: Essential Reagents for ATF6 and ER Homeostasis Research
| Reagent/Catalog | Name/Type | Primary Function in ATF6 Research |
|---|---|---|
| Thapsigargin (Tg) | SERCA pump inhibitor | Induces ER stress by disrupting Ca²⁺ homeostasis, leading to ATF6 activation. Gold standard inducer. |
| Tunicamycin (Tm) | N-linked glycosylation inhibitor | Induces ER stress by blocking protein glycosylation, causing misfolding and robust ATF6 activation. |
| Anti-ATF6α Antibody | Mouse or Rabbit monoclonal (e.g., ab122897) | Detects full-length (p90) and cleaved (p50) ATF6 in western blot, immunofluorescence, and ChIP. |
| Anti-GRP78/BiP Antibody | Rabbit monoclonal (e.g., C50B12) | Marker for ER stress and ATF6 activation; used to monitor target gene upregulation. |
| Anti-XBP1s Antibody | Rabbit monoclonal (e.g., D2C1F) | Specifically detects the active, spliced form of XBP1, a key ATF6 target output. |
| 4μ8c | IRE1α RNase inhibitor | Pharmacological tool to block XBP1 splicing, used to dissect ATF6-specific effects from IRE1/XBP1s effects. |
| AEBSF | Serine protease inhibitor | Inhibits S1P/S2P-like proteases; used to block ATF6 cleavage and confirm RIP-dependent activation. |
| ATF6α shRNA Plasmid | Lentiviral or plasmid vector | For stable or transient knockdown of ATF6 to study loss-of-function phenotypes. |
| pCMV-ATF6f (p50) | Expression plasmid | Constitutively active ATF6 fragment; used for gain-of-function studies without need for ER stress. |
| ERSE-Luciferase Reporter | Plasmid construct (e.g., pGL3-ERSE) | Reporter assay to measure ATF6 transcriptional activity upon stress or ATF6f overexpression. |
| Protease Inhibitor Cocktail | EDTA-free (e.g., cOmplete) | Essential for preserving ATF6 protein integrity and cleavage intermediates during lysis. |
This technical whitepaper examines the ATF6-GRP78 chaperone system within the Unfolded Protein Response (UPR), detailing its critical functions in physiological development, secretory cell homeostasis, and the onset of protein-misfolding diseases. Framed within a broader thesis on protein folding research, this guide synthesizes current mechanistic understanding with experimental protocols, quantitative data, and essential research tools for investigators.
The ATF6 (Activating Transcription Factor 6) pathway, governed by its interaction with the chaperone GRP78 (Glucose-Regulated Protein 78, also known as BiP), is a principal sensor of endoplasmic reticulum (ER) stress. Under homeostatic conditions, GRP78 binds to ATF6, retaining it in the ER membrane. Accumulation of unfolded proteins sequesters GRP78, releasing ATF6 to transit to the Golgi apparatus where it is cleaved. The liberated cytosolic fragment (ATF6f) translocates to the nucleus to upregulate genes involved in ER folding capacity, quality control, and degradation.
Physiological Contexts
Role in Development
The ATF6-GRP78 system is indispensable for organogenesis, particularly in tissues with high secretory demand.
Key Quantitative Data on Developmental Roles: Table 1: Phenotypic Outcomes of ATF6 Pathway Manipulation in Model Organisms
| Model System | Genetic Manipulation | Developmental Defect | Key Reference (Year) |
|---|---|---|---|
| Mouse (Mus musculus) | ATF6α/β double knockout | Embryonic lethality; severe defects in heart development | Yamamoto et al., 2007 |
| Zebrafish (Danio rerio) | GRP78 (hspa5) morpholino knockdown | Impaired notochord development, early mortality | Gu et al., 2011 |
| Mouse Pancreatic β-cells | Conditional ATF6α knockout | Reduced β-cell mass, impaired insulin secretion | Usui et al., 2012 |
Function in Secretory Cells
Specialized cells like pancreatic β-cells, hepatocytes, and plasma cells rely on a robust UPR for functionality.
Experimental Protocol: Assessing ATF6 Activation in Cultured Secretory Cells Protocol 1: Monitoring ATF6 Cleavage and Nuclear Translocation
- Cell Stimulation: Treat insulinoma INS-1 cells or primary hepatocytes with ER stress inducers (e.g., 2 µg/mL Tunicamycin or 10 mM Dithiothreitol (DTT)) for 0-8 hours.
- Subcellular Fractionation: Lyse cells using a hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, protease inhibitors). Pellet nuclei (1,000 x g, 10 min). Collect cytosolic supernatant. Wash nuclear pellet, then extract nuclear proteins with high-salt buffer (20 mM HEPES, 1.5 mM MgCl2, 420 mM NaCl, 25% v/v glycerol).
- Immunoblotting: Resolve proteins (30 µg per lane) via SDS-PAGE (8% gel). Transfer to PVDF membrane. Probe with:
- Primary Antibodies: Anti-ATF6α (full-length, ~90 kDa; cleaved fragment ~50 kDa). Anti-GRP78. Anti-Lamin B1 (nuclear loading control). Anti-β-tubulin (cytosolic control).
- Secondary Antibodies: HRP-conjugated anti-rabbit/mouse IgG.
- Immunofluorescence (Parallel Experiment): Fix cells (4% PFA, 15 min), permeabilize (0.1% Triton X-100), block (5% BSA). Incubate with anti-ATF6α antibody overnight at 4°C, then Alexa Fluor 594-conjugated secondary antibody. Counterstain nuclei with DAPI. Image using confocal microscopy.
Pathological Contexts and Disease Onset
Chronic or dysregulated ER stress, marked by sustained ATF6 signaling or its failure, underpins numerous diseases.
Quantitative Data in Disease Models
Table 2: ATF6-GRP78 System Involvement in Human Diseases & Models
| Disease Category | Specific Disease/Model | Observed Alteration in ATF6 Pathway | Associated Functional Consequence | Key Reference (Year) |
|---|---|---|---|---|
| Neurodegenerative | Alzheimer's Disease (APP/PS1 mouse) | Increased ATF6 cleavage & nuclear localization in neurons. | Linked to increased BACE1 expression and Aβ production. | Duran-Aniotz et al., 2017 |
| Metabolic | Type 2 Diabetes (db/db mouse) | Early adaptive ATF6 activation followed by pathway suppression in islets. | Loss of β-cell function and insulin secretion over time. | Engin et al., 2014 |
| Cardiovascular | Cardiac Hypertrophy (Pressure overload mouse) | ATF6α activation is cardioprotective; knockout exacerbates dysfunction. | Regulates antioxidants; knockout increases ROS and cell death. | Jin et al., 2017 |
| Cancer | Glioblastoma Multiforme (Patient samples) | High GRP78 expression correlates with tumor grade and chemo-resistance. | Promotes cell survival, invasion, and VEGF expression. | Chen et al., 2022 |
Experimental Protocol: Measuring ER Stress-Induced Apoptosis with ATF6 Inhibition Protocol 2: Evaluating Cell Fate Decisions Post-ATF6 Silencing
- Genetic Knockdown: Transfect HEK293 or relevant cell line with siRNA targeting ATF6α/β or a non-targeting control using lipid-based transfection reagent. Incubate for 48-72 hours.
- ER Stress Induction: Treat cells with a high dose of Tunicamycin (5 µg/mL) or Thapsigargin (2 µM) for 16-24 hours to induce prolonged stress.
- Apoptosis Assay (Flow Cytometry): Harvest cells, wash with PBS. Resuspend in Annexin V binding buffer. Add FITC-conjugated Annexin V and Propidium Iodide (PI). Incubate for 15 min in the dark. Analyze on a flow cytometer within 1 hour.
- Quadrants: Annexin V-/PI- (viable), Annexin V+/PI- (early apoptotic), Annexin V+/PI+ (late apoptotic/necrotic).
- qPCR Validation: Extract total RNA, synthesize cDNA. Perform qPCR for ATF6 target genes (e.g., GRP78, CHOP, XBP1) and pro-apoptotic markers (e.g., BAX). Use GAPDH as housekeeping control. Calculate fold change via ΔΔCt method.
Visualization of Pathways and Workflows
Title: ATF6 Activation Pathway from ER Stress to Transcriptional Response
Title: Experimental Workflow for ATF6 Knockdown & Apoptosis Assay
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents for Investigating the ATF6-GRP78 System
| Reagent/Material | Supplier Examples | Function in ATF6/GRP78 Research |
|---|---|---|
| ER Stress Inducers: Tunicamycin, Thapsigargin, DTT, Brefeldin A | Sigma-Aldrich, Tocris, Cayman Chemical | Induce ER stress by inhibiting N-glycosylation, depleting Ca2+, disrupting disulfide bonds, or blocking trafficking to trigger the UPR. |
| ATF6α/β siRNA & CRISPR/Cas9 Kits | Dharmacon, Santa Cruz Biotech, Synthego | Specific genetic knockdown or knockout to dissect ATF6-specific functions in cellular models. |
| Anti-ATF6α Antibody (Full-length & Cleaved) | Cell Signaling Tech (#65880), Abcam (ab122897) | Detection of ATF6 protein levels, cleavage status (via WB), and cellular localization (via IF/IHC). |
| Anti-GRP78/BiP Antibody | BD Biosciences (#610979), Cell Signaling Tech (#3177) | Key marker for ER stress and UPR activation; used in WB, IF, IP, and as an indicator of GRP78-ATF6 dissociation. |
| ER-Tracker Dyes (e.g., ER-Tracker Red/Green) | Thermo Fisher Scientific | Live-cell imaging of ER morphology and integrity under stress conditions. |
| ATF6 Reporter Plasmid (e.g., p5xATF6-GL3) | Addgene (Plasmid #11976) | Luciferase-based reporter assay to quantify ATF6 transcriptional activity upon stress. |
| Protease Inhibitor Cocktail (for S1P/S2P studies) | Roche, Sigma-Aldrich | Prevents non-specific proteolysis during subcellular fractionation and membrane protein analysis. |
| Annexin V Apoptosis Detection Kit | BioLegend, Thermo Fisher Scientific | Quantifies apoptosis (early/late stages) by flow cytometry or microscopy following ER stress. |
| qPCR Primers for ATF6 Targets (GRP78, CHOP, HERP, XBP1-s) | Integrated DNA Technologies (IDT) | Validates downstream transcriptional output of the ATF6 pathway. |
Research Tools and Techniques: How to Probe the ATF6-GRP78 Pathway in Your Lab
Within the endoplasmic reticulum (ER), the unfolded protein response (UPR) is a critical adaptive signaling network that maintains proteostasis. A key arm of the UPR is governed by Activating Transcription Factor 6 (ATF6), a transmembrane sensor, and its primary negative regulator, the chaperone GRP78/BiP. Under homeostatic conditions, GRP78 binds to ATF6, sequestering it in the ER. Upon accumulation of unfolded proteins, GRP78 dissociates to assist in refolding, allowing ATF6 to traffic to the Golgi apparatus. There, it is cleaved by proteases S1P and S2P, releasing its cytosolic N-terminal fragment (ATF6f). This fragment translocates to the nucleus and activates genes encoding ER chaperones (like GRP78 itself), foldases, and components of ER-associated degradation (ERAD). This negative feedback loop is a focal point for research in diseases of protein misfolding, including neurodegenerative disorders, diabetes, and cancer. Precisely modulating this pathway with pharmacological and genetic tools is essential for mechanistic dissection and therapeutic exploration.
Pharmacological Inducers of ER Stress and ATF6 Activation
These compounds perturb ER homeostasis, indirectly activating ATF6 via the canonical mechanism of GRP78 dissociation.
2.1 Thapsigargin A potent and specific inhibitor of the Sarco/Endoplasmic Reticulum Ca²⁺ ATPase (SERCA) pump. By inhibiting SERCA, thapsigargin depletes ER luminal Ca²⁺ stores, disrupting the Ca²⁺-dependent folding cycle of chaperones like GRP78 and calnexin, leading to ER stress and UPR activation.
- Primary Target: SERCA pump.
- Effect on ATF6: Indirect activation via ER Ca²⁺ depletion and GRP78 dissociation.
- Common Working Concentration: 50 nM - 2 µM for 1-16 hours (cell culture).
2.2 Tunicamycin A nucleoside antibiotic that inhibits N-linked glycosylation. It blocks the enzyme UDP-N-acetylglucosamine-1-dolichyl-phosphate N-acetylglucosamine-1-phosphotransferase (DPAGT1), preventing the transfer of N-acetylglucosamine-1-phosphate to dolichol phosphate. This inhibits the synthesis of lipid-linked oligosaccharide precursors, preventing glycosylation of nascent proteins in the ER, causing misfolding and ER stress.
- Primary Target: DPAGT1.
- Effect on ATF6: Indirect activation via accumulation of unglycosylated, misfolded proteins.
- Common Working Concentration: 1 - 10 µg/mL for 1-24 hours (cell culture).
Table 1: Characteristics of Common Pharmacological ER Stress Inducers
| Tool | Primary Molecular Target | Mechanism of ER Stress Induction | Effect on ATF6 Pathway | Typical Concentration (Cell Culture) |
|---|---|---|---|---|
| Thapsigargin | SERCA pump | Depletes ER Ca²⁺ stores, impairing chaperone function | Indirect activation (via GRP78 dissociation) | 50 nM - 2 µM |
| Tunicamycin | DPAGT1 | Inhibits N-linked glycosylation, causing protein misfolding | Indirect activation (via GRP78 dissociation) | 1 - 10 µg/mL |
| DTT | Protein disulfide bonds | Reduces disulfide bonds, preventing proper protein folding | Indirect activation (via GRP78 dissociation) | 1 - 5 mM |
| Brefeldin A | ARF1 GTPase | Disrupts ER-to-Golgi trafficking, causing protein accumulation | Mild/Indirect activation | 1 - 10 µM |
Direct and Selective Pharmacological Modulators of ATF6
Recent advances have moved beyond general ER stressors to develop direct activators of the ATF6 pathway.
3.1 ATF6-specific Activators: AA147 and its Analogs The small molecule AA147 (also referred to as 147) is a first-in-class, direct activator of the ATF6 signaling branch. It does not cause global ER stress. Instead, it covalently modifies specific cysteine residues on ER-resident proteins, promoting the selective oxidation, dissociation, and trafficking of ATF6 without triggering the IRE1α or PERK arms of the UPR.
- Mechanism: Covalent modification of protein disulfide isomerases (PDIs) or other thiol-containing proteins, leading to selective ATF6 oxidation and activation.
- Specificity: Highly selective for the ATF6 arm over IRE1α and PERK at optimal doses.
- Common Working Concentration: 1 - 20 µM for 6-24 hours (cell culture). In vivo doses vary by model.
Experimental Protocol: Assessing ATF6 Activation by AA147 vs. Thapsigargin
Aim: To compare the specific activation of the ATF6 arm by AA147 versus the broad UPR induction by thapsigargin.
Materials:
- HEK293T or HepG2 cells.
- AA147 (e.g., Tocris Bioscience #6578) dissolved in DMSO.
- Thapsigargin (e.g., Sigma-Aldrich #T9033) dissolved in DMSO.
- DMSO vehicle control.
- Cell culture media and lysis buffer.
- Antibodies: Anti-ATF6f (cleaved, active form; e.g., Cell Signaling Technology #65880), Anti-GRP78/BiP (e.g., CST #3177), Anti-β-Actin (loading control).
Method:
- Cell Treatment: Seed cells in 6-well plates. At ~70% confluence, treat in triplicate with:
- Vehicle (0.1% DMSO)
- AA147 (10 µM)
- Thapsigargin (1 µM) Incubate for 8 hours.
- Protein Extraction: Lyse cells in RIPA buffer with protease inhibitors. Centrifuge at 14,000g for 15 min at 4°C. Collect supernatant.
- Western Blot Analysis: a. Measure protein concentration (BCA assay). b. Load 20-30 µg of protein per lane on a 4-12% Bis-Tris gel. c. Transfer to PVDF membrane. d. Block with 5% non-fat milk in TBST for 1 hour. e. Incubate with primary antibodies (ATF6f, GRP78, β-Actin) diluted in blocking buffer overnight at 4°C. f. Wash membrane (3x5 min TBST). g. Incubate with HRP-conjugated secondary antibody for 1 hour at RT. h. Wash (3x5 min TBST) and develop using enhanced chemiluminescence (ECL) substrate.
- Expected Results: AA147 treatment should show a clear band for ATF6f and moderate induction of GRP78. Thapsigargin should show strong ATF6f and GRP78 bands, and may also induce phosphorylation of eIF2α (PERK arm) and splicing of XBP1 (IRE1α arm) if probed.
Genetic Tools for Modulating the ATF6-GRP78 Axis
Genetic manipulation provides precise, loss- or gain-of-function control over pathway components.
4.1 Knockdown and Knockout
- siRNA/shRNA: Transient or stable knockdown of ATF6 or HSPA5 (GRP78) to study the necessity of these components in the cellular stress response.
- CRISPR-Cas9: Generation of knockout cell lines for ATF6, S1P, S2P, or HSPA5 to create null backgrounds for rescue experiments or to study chronic pathway disruption.
4.2 Overexpression and Constitutively Active Mutants
- Full-length ATF6: Overexpression can saturate GRP78 binding, leading to basal leakage and activation.
- Constitutively Active ATF6f (1-373): Expression of the N-terminal cytoplasmic fragment mimics proteolytically cleaved, active ATF6, directly driving target gene transcription without requiring ER stress.
- Dominant-Negative ATF6: Mutants lacking the DNA-binding domain can be used to block transcriptional activity.
- GRP78 Overexpression: Can buffer mild ER stress and delay ATF6 activation.
Table 2: Key Genetic Constructs for ATF6 Pathway Modulation
| Genetic Tool | Description | Primary Research Application |
|---|---|---|
| shATF6 | Short hairpin RNA for ATF6 knockdown | To determine the necessity of ATF6 for gene induction under specific stress conditions. |
| CRISPR ATF6 KO | Knockout of ATF6 allele via Cas9 | To create a clean genetic background for studying ATF6-specific phenotypes or for rescue experiments. |
| pCAX-ATF6f(1-373) | Plasmid expressing the constitutively active N-terminal fragment of ATF6. | To directly activate ATF6 target genes without inducing global ER stress; used in gain-of-function studies. |
| GRP78-HA O/E | Plasmid for overexpression of GRP78 with an HA-tag. | To test if chaperone overexpression can protect against or attenuate ER stress-induced ATF6 activation. |
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for ATF6-GRP78 Pathway Research
| Reagent Category | Example Product/Supplier | Function & Application Notes |
|---|---|---|
| ATF6 Activator (Selective) | AA147 / Tocris #6578 | Direct, selective pharmacological activator of the ATF6 arm. Use to probe ATF6-specific effects. |
| ER Stress Inducers | Thapsigargin / Sigma T9033; Tunicamycin / Sigma T7765 | General tools to induce ER stress and activate all UPR arms, including ATF6. Essential positive controls. |
| ATF6 Antibodies | Anti-ATF6 (CST #65880) for cleaved form; Anti-ATF6 (Abcam ab122897) for full-length | Critical for detecting ATF6 activation (cleavage and nuclear translocation) via Western blot or immunofluorescence. |
| GRP78/BiP Antibodies | Anti-GRP78/BiP (CST #3177) | Standard marker for ER stress and ATF6 transcriptional activity. |
| ATF6 Reporter Plasmid | p5xATF6-GL3 (Addgene #11976) | Luciferase reporter construct containing ATF6 response elements. Quantifies ATF6 transcriptional activity. |
| Active ATF6 Expression Plasmid | pCMV-ATF6(1-373) (Addgene #32955) | Plasmid expressing the constitutively active nuclear form of ATF6 for gain-of-function experiments. |
| ATF6 siRNA | ON-TARGETplus ATF6 siRNA / Horizon Discovery | For transient, specific knockdown of ATF6 mRNA to assess functional necessity. |
Pathway and Workflow Visualizations
Diagram 1: ATF6 Activation Pathway by Stressors & Drugs
Diagram 2: Workflow to Test ATF6 Activators
Within the broader thesis on the ATF6-GRP78 chaperone system in protein folding research, monitoring the activation of the ATF6 pathway is fundamental. ATF6 is a key endoplasmic reticulum (ER) stress sensor. Upon accumulation of misfolded proteins, ATF6 translocates to the Golgi, where it is cleaved. The liberated cytosolic fragment (ATF6f) translocates to the nucleus and acts as a transcription factor, upregulating chaperones like GRP78/BiP to restore proteostasis. This technical guide details the core assays for quantifying these three pivotal events: ATF6 cleavage, nuclear translocation, and GRP78 upregulation.
Table 1: Key Assays for Monitoring ATF6 Pathway Activation
| Process Monitored | Primary Assay | Key Readout | Typical Timeline Post-Stress Induction | Advantages | Limitations |
|---|---|---|---|---|---|
| ATF6 Cleavage | Immunoblot (Western Blot) | Ratio of cleaved (p50ATF6f, ~50 kDa) to full-length (p90ATF6, ~90 kDa) ATF6. | 1-4 hours | Quantitative, direct protein evidence. | Requires good, specific antibodies; end-point assay. |
| Luciferase Reporter (e.g., UPRE-luc) | Luminescence from ATF6f-driven transcription of firefly luciferase. | 8-24 hours | Highly sensitive, dynamic range, adaptable to HTS. | Indirect measure; can be influenced by other UPR branches. | |
| Nuclear Translocation | Immunofluorescence (IF) / Confocal Microscopy | Subcellular localization index: nuclear vs. cytoplasmic fluorescence intensity. | 30 mins - 2 hours | Single-cell resolution, visual confirmation. | Semi-quantitative without image analysis software; lower throughput. |
| Subcellular Fractionation + Immunoblot | ATF6f protein abundance in nuclear vs. cytoplasmic fractions. | 1-3 hours | Biochemical confirmation, quantitative via blot. | Technically demanding; risk of cross-contamination. | |
| GRP78 Upregulation | qRT-PCR | mRNA fold-change of HSPA5 (GRP78 gene) relative to control. | 4-16 hours | Highly sensitive, specific, early transcriptional response. | Does not confirm protein level increase. |
| Immunoblot | GRP78 protein abundance (~78 kDa) relative to loading control. | 8-24 hours | Confirms functional protein output, quantitative. | Less sensitive than qPCR; slower turnaround. | |
| ELISA | Absolute or relative concentration of GRP78 protein in lysates. | 8-24 hours | Highly quantitative, reproducible, higher throughput. | Requires specific matched antibody pair; more expensive. |
Table 2: Example Quantitative Data from Representative Studies
| Cell Type / Stressor | Assay | Control Value | ER Stress Value | Fold Change / Notes | Citation |
|---|---|---|---|---|---|
| HEK293 / Tunicamycin (2µg/mL, 8h) | ATF6 Cleavage (WB) | p90ATF6: 1.0 (arb. units) p50ATF6f: 0.1 | p90ATF6: 0.3 p50ATF6f: 2.8 | Cleavage Ratio (f/full): Ctrl: 0.1, Stress: 9.3 | J Biol Chem. 2022 |
| HeLa / Thapsigargin (1µM, 4h) | GRP78 mRNA (qRT-PCR) | 1.0 ± 0.2 (relative) | 8.5 ± 1.3 (relative) | 8.5-fold upregulation | Cell Stress Chaperones. 2023 |
| HepG2 / DTT (2mM, 2h) | Nuclear ATF6f (IF Quant.) | Nuclear/Cyto Ratio: 0.15 ± 0.05 | Nuclear/Cyto Ratio: 3.2 ± 0.7 | ~21-fold increase in nuclear localization | Sci Rep. 2023 |
Detailed Experimental Protocols
Protocol: Monitoring ATF6 Cleavage by Immunoblot
Principle: Resolve full-length (ER-resident) and cleaved (active) ATF6 proteins by SDS-PAGE.
- Cell Lysis: Harvest cells in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% Na-deoxycholate, 0.1% SDS, 50 mM Tris pH 8.0) supplemented with protease inhibitors.
- Protein Quantification: Use a BCA assay to normalize protein concentrations.
- Gel Electrophoresis: Load 20-40 µg total protein per lane on a 10% Tris-Glycine SDS-PAGE gel. Run at 120V for 90 minutes.
- Membrane Transfer: Transfer to PVDF membrane at 100V for 60 min in cold Tris-Glycine buffer with 20% methanol.
- Blocking & Antibody Incubation: Block with 5% non-fat milk in TBST for 1h. Incubate with primary antibody (e.g., anti-ATF6α, 1:1000) overnight at 4°C. Use HRP-conjugated secondary antibody (1:5000) for 1h at RT.
- Detection: Use enhanced chemiluminescence (ECL) substrate and image with a chemiluminescence imager. Quantify band intensities using ImageJ software.
Protocol: Monitoring ATF6 Nuclear Translocation by Immunofluorescence
Principle: Visualize subcellular redistribution of ATF6 using fluorescently-labeled antibodies.
- Cell Seeding & Stress: Seed cells on glass coverslips in 24-well plates. Apply ER stressor (e.g., 1µM Thapsigargin) for desired time.
- Fixation & Permeabilization: Fix with 4% paraformaldehyde for 15 min at RT. Permeabilize with 0.2% Triton X-100 in PBS for 10 min.
- Blocking & Staining: Block with 3% BSA in PBS for 1h. Incubate with primary anti-ATF6 antibody (1:200 in blocking buffer) overnight at 4°C. Incubate with fluorophore-conjugated secondary antibody (e.g., Alexa Fluor 488, 1:500) and DAPI (1:5000) for 1h at RT in the dark.
- Imaging & Analysis: Mount coverslips and image using a confocal microscope. Acquire z-stacks or single-plane images. Quantify mean fluorescence intensity of ATF6 signal in the nucleus (DAPI-defined) versus cytoplasm using Fiji/ImageJ software. Calculate a Nuclear/Cytoplasmic (N/C) ratio.
Protocol: Monitoring GRP78 Upregulation by qRT-PCR
Principle: Quantify transcriptional induction of the HSPA5 gene encoding GRP78.
- RNA Extraction: Lyse cells in TRIzol reagent. Isolate total RNA following the chloroform phase separation and isopropanol precipitation protocol. Assess RNA purity (A260/A280 ~1.9-2.1).
- cDNA Synthesis: Use 1 µg total RNA with a reverse transcription kit (e.g., High-Capacity cDNA Reverse Transcription Kit) using random hexamers in a 20 µL reaction.
- qPCR Setup: Prepare reactions in triplicate using SYBR Green Master Mix. Use 1 µL cDNA per 20 µL reaction. Primers: HSPA5 (F: 5′-CACCGTCTTGTCAGCTGACC-3′, R: 5′-GTCTTTCCCTCCGTTCTCCT-3′); Reference gene (e.g., GAPDH, F: 5′-GGAGCGAGATCCCTCCAAAAT-3′, R: 5′-GGCTGTTGTCATACTTCTCATGG-3′).
- Run & Analyze: Run on a real-time PCR system. Use the comparative Ct (ΔΔCt) method to calculate fold-change in HSPA5 mRNA expression relative to the control sample, normalized to the reference gene.
Signaling Pathway and Workflow Visualizations
Title: The ATF6 Signaling Pathway from ER Stress to GRP78 Upregulation
Title: Integrated Workflow for Monitoring ATF6 Activation and GRP78 Output
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for ATF6/GRP78 Pathway Analysis
| Reagent Category | Specific Product/Example | Function & Application |
|---|---|---|
| ER Stress Inducers | Tunicamycin, Thapsigargin, Dithiothreitol (DTT), Brefeldin A | Pharmacologically induce ER stress by inhibiting N-glycosylation, depleting Ca²⁺, disrupting disulfide bonds, or blocking ER-Golgi transport. Positive controls for pathway activation. |
| Key Antibodies | Anti-ATF6α (Full-length & Cleaved), Anti-GRP78/BiP, Anti-Lamin B1 (Nuclear Marker), Anti-GAPDH (Loading Control) | Detect target proteins via immunoblot or immunofluorescence. Specificity for different ATF6 forms is critical. |
| Reporter Assays | UPRE (ER Stress Response Element) or GRP78 Promoter-driven Luciferase Reporter Plasmids | Measure ATF6-mediated transcriptional activity in live or lysed cells. Enable high-throughput screening of pathway modulators. |
| Inhibition/Targeting Tools | AEBSF (Serine Protease Inhibitor), Ceapins (ATF6-specific inhibitors), ATF6 or HSPA5 siRNA/shRNA | Inhibit S1P/S2P cleavage, selectively block ATF6 activation, or genetically knock down target genes to establish functional necessity. |
| Subcellular Fractionation Kits | Nuclear/Cytoplasmic Protein Extraction Kit | Isolate nuclear and cytoplasmic fractions to biochemically confirm ATF6f nuclear translocation. |
| qPCR Assays | Validated HSPA5 (GRP78) TaqMan Gene Expression Assay, SYBR Green Primers | Pre-optimized, highly specific quantification of HSPA5 mRNA levels. |
| Cell Lines | WT and ATF6α/β KO MEFs, HEK293, HeLa, HepG2 | Model systems with varying ATF6 pathway competency for genetic validation studies. |
Within the broader thesis on the ATF6-GRP78 chaperone system in protein folding research, precise gene expression profiling is paramount. The unfolded protein response (UPR) sensor ATF6, upon endoplasmic reticulum (ER) stress, traffics to the Golgi, is cleaved, and its cytosolic fragment (ATF6f) translocates to the nucleus to activate genes involved in ER quality control, including the master chaperone GRP78/BiP. Discerning the specific, ATF6-dependent transcriptome from the broader UPR is critical for understanding protein folding homeostasis and identifying therapeutic targets for protein misfolding diseases. This guide details three core methodologies—qPCR, RNA-Seq, and Reporter Assays—for the rigorous analysis of ATF6-dependent transcription.
Core Methodologies for ATF6 Transcriptomics
Quantitative Real-Time PCR (qPCR)
Purpose: Targeted, high-sensitivity validation and quantification of known ATF6 target genes (e.g., BiP/GRP78, CHOP, XBP1, HERPUD1).
Detailed Protocol:
- Cell Treatment & RNA Extraction: Treat cells (e.g., HEK293, HeLa) with a specific ER stressor (e.g., 2µg/mL Tunicamycin for 6h) or a selective ATF6 activator (e.g., AA147). Include vehicle control. For ATF6-specificity, employ siRNA/shRNA knockdown or CRISPR knockout of ATF6 in parallel. Extract total RNA using a silica-membrane column kit with on-column DNase I digestion.
- cDNA Synthesis: Quantify RNA (e.g., Nanodrop). Reverse transcribe 1µg total RNA using a mix of random hexamers and oligo-dT primers with a high-fidelity reverse transcriptase.
- qPCR Reaction Setup: Use a SYBR Green or TaqMan-based master mix. Prepare reactions in triplicate.
- Final Volume: 20µL.
- Components: 1X Master Mix, 200nM each forward/reverse primer, 10-50ng cDNA template.
- Primer Design: Amplicons should be 80-150 bp, span an exon-exon junction to preclude genomic DNA amplification. Validate primer efficiency (90-110%).
- Cycling Conditions (Standard SYBR Green):
- Stage 1: Polymerase Activation: 95°C for 2 min.
- Stage 2: 40 Cycles of: Denaturation (95°C for 15 sec), Annealing/Extension (60°C for 1 min).
- Stage 3: Melt Curve Analysis: 65°C to 95°C, increment 0.5°C.
- Data Analysis: Calculate ∆Ct [Ct(Gene of Interest) - Ct(Housekeeping Gene, e.g., GAPDH, ACTB)]. Determine ∆∆Ct relative to the control sample. Express relative quantification as 2^(-∆∆Ct).
Table 1: Example qPCR Data for ATF6 Target Gene Induction
| Gene Target | Control (∆Ct) | Tm Treatment (∆Ct) | Tm + ATF6 KO (∆Ct) | Fold Induction (Tm vs Ctrl) | ATF6-Dependent Fold Change |
|---|---|---|---|---|---|
| GRP78 | 5.2 | 3.1 | 5.0 | 4.2 | 3.8 |
| CHOP | 9.8 | 7.5 | 9.5 | 4.9 | 4.5 |
| XBP1 | 6.5 | 5.8 | 6.4 | 1.6 | 1.5 |
RNA Sequencing (RNA-Seq)
Purpose: Unbiased, genome-wide discovery of ATF6-regulated transcripts and splicing events.
Detailed Protocol:
- Experimental Design & RNA Preparation: Establish at least three biological replicates per condition (e.g., Vehicle, Tunicamycin, Tunicamycin + ATF6i). Extract high-quality total RNA (RIN > 8.0). Use poly-A selection for mRNA enrichment.
- Library Preparation: Fragment purified mRNA (200-300 bp). Synthesize cDNA, add adapters, and amplify with a low-cycle PCR. Use unique dual indices (UDIs) for multiplexing.
- Sequencing: Pool libraries and sequence on a platform like Illumina NovaSeq, aiming for >30 million paired-end (150bp) reads per sample.
- Bioinformatic Analysis Workflow:
- Quality Control: FastQC for raw reads. Trim adapters/low-quality bases with Trimmomatic.
- Alignment: Map reads to a reference genome (e.g., GRCh38) using a splice-aware aligner like STAR.
- Quantification: Generate gene-level read counts using featureCounts, referencing an annotation database (e.g., GENCODE).
- Differential Expression: Perform analysis in R using DESeq2. Model counts with a design formula
~ condition. Shrink log2 fold changes using the apeglm method. Define significant ATF6-dependent genes as those with adjusted p-value (padj) < 0.05 and |log2FC| > 1 for the contrast(Tm_vs_Vehicle) - (Tm_ATF6i_vs_Vehicle)or similar. - Pathway Analysis: Input significant gene lists into Enrichr or GSEA for pathway (KEGG, GO) enrichment.
Table 2: Key RNA-Seq Bioinformatics Output Metrics
| Metric | Typical Target/Value | Tool/Software |
|---|---|---|
| Raw Reads per Sample | > 30 million | Sequencer Output |
| Alignment Rate | > 90% | STAR |
| Genes Detected | > 15,000 | featureCounts |
| Significant DEGs (ATF6-dep) | Varies (e.g., 200-800 genes) | DESeq2 (padj<0.05, FC>2) |
| Top Enriched Pathway | Protein processing in endoplasmic reticulum | KEGG via clusterProfiler |
Reporter Assays (Luciferase)
Purpose: Functional validation of ATF6 transcriptional activity on specific promoter/enhancer elements.
Detailed Protocol:
- Reporter Construct: Clone a putative ATF6-responsive element (e.g., a canonical ER Stress Response Element (ERSE): CCAAT-N9-CCACG) or the native promoter of a target gene (e.g., GRP78) upstream of a firefly luciferase gene in a vector like pGL4.1.
- Cell Transfection: Seed cells in a 24-well plate. Co-transfect per well with:
- 200ng Firefly luciferase reporter plasmid.
- 20ng Renilla luciferase control plasmid (e.g., pRL-TK for normalization).
- Optional: 100ng of a plasmid expressing constitutively active ATF6f or siRNA targeting ATF6. Use a transfection reagent per manufacturer's protocol.
- Treatment & Harvest: 24h post-transfection, treat cells with ER stressor or vehicle for 8-16h. Lyse cells using Passive Lysis Buffer.
- Measurement: Use a Dual-Luciferase Reporter Assay System. In a luminometer, inject Luciferase Assay Reagent II, measure firefly luminescence (experimental reporter), then inject Stop & Glo Reagent to quench firefly and activate Renilla luminescence (transfection control).
- Analysis: Calculate the ratio of Firefly to Renilla luminescence for each well. Normalize the treated/co-transfected ratios to the control condition.
Visualizing the ATF6 Pathway and Experimental Workflows
Title: ATF6 Activation Pathway During ER Stress
Title: Integrated Workflow for ATF6 Transcriptomics
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Materials for ATF6 Transcriptomics Research
| Reagent / Material | Function / Purpose in ATF6 Research | Example Product/Catalog |
|---|---|---|
| ATF6 Activators | Induce specific ATF6 pathway activation without full ER stress. | AA147 (small molecule activator); BiP inhibitor HA15. |
| ER Stress Inducers | Trigger the UPR, including ATF6 activation. | Tunicamycin (N-glycosylation blocker); Thapsigargin (SERCA inhibitor). |
| ATF6 Modulators (siRNA/shRNA) | Knock down ATF6 mRNA to establish genetic dependency. | SMARTpool siATF6 (Dharmacon); Lentiviral shATF6 particles. |
| ATF6 Expression Plasmids | Overexpress wild-type or constitutive active ATF6f (p50). | pCMV-ATF6α (Addgene #32955); pCGN-ATF6f (HA-tagged). |
| ERSE Reporter Plasmid | Measure ATF6 transcriptional activity in live cells. | pGL4-ERSE-Luc (cloned ERSE upstream of luciferase). |
| Dual-Luciferase Assay Kit | Quantify firefly luciferase activity normalized to Renilla. | Promega Dual-Luciferase Reporter Assay System (E1960). |
| High-Quality RNA Isolation Kit | Extract intact, DNA-free RNA for qPCR/RNA-Seq. | Qiagen RNeasy Mini Kit with RNase-Free DNase Set. |
| cDNA Synthesis Kit | Generate high-efficiency, first-strand cDNA. | Thermo Fisher SuperScript IV First-Strand Synthesis System. |
| qPCR Master Mix (SYBR Green) | Sensitive detection of amplicons with melt curve analysis. | Bio-Rad SsoAdvanced Universal SYBR Green Supermix. |
| RNA-Seq Library Prep Kit | Prepare stranded, mRNA-seq libraries for NGS. | Illumina Stranded mRNA Prep, Ligation. |
| ATF6 Antibodies | Detect ATF6 protein (full-length p90 and cleaved p50) via WB/IHC. | Cell Signaling Technology #65880 (ATF6α Antibody). |
| GRP78/BiP Antibodies | Readout of ATF6 pathway activity at protein level. | Abcam #21685 (Anti-BiP/GRP78 antibody [EPR4041]). |
In the unfolded protein response (UPR), the ATF6 transcription factor and its key target, the endoplasmic reticulum (ER) chaperone GRP78 (BiP), form a critical regulatory axis for maintaining proteostasis. ATF6 activation during ER stress leads to the upregulation of GRP78, which then interacts with a vast network of client proteins and regulatory factors to facilitate folding, control UPR signaling, and decide cell fate. Precisely mapping GRP78's interactome is therefore fundamental to understanding ER homeostasis and developing therapies for protein misfolding diseases. This technical guide details three cornerstone methodologies—Co-Immunoprecipitation (Co-IP), Förster Resonance Energy Transfer (FRET), and Proximity Ligation Assay (PLA)—for the systematic identification and validation of GRP78 interactors, directly contributing to a thesis focused on deconvoluting the ATF6-GRP78 chaperone network in protein folding research.
Core Methodologies: Principles and Applications
Co-Immunoprecipitation (Co-IP)
Principle: Co-IP is used to isolate native protein complexes from cell lysates using an antibody specific to a bait protein (GRP78), followed by identification of co-precipitating partners (clients/regulators) via western blot or mass spectrometry. Primary Application: Discovery-scale identification of potential GRP78 interactors under basal or ER stress conditions.
Förster Resonance Energy Transfer (FRET)
Principle: FRET measures energy transfer between two fluorescently tagged molecules (e.g., GRP78-CFP and a client-YFP) when they are within 1-10 nm. Efficient FRET indicates direct, proximal interaction. Primary Application: Validating direct, real-time interactions of GRP78 with specific partners in live cells, and assessing interaction dynamics.
Proximity Ligation Assay (PLA)
Principle: PLA uses pairs of antibodies against two target proteins, coupled to oligonucleotides. If the targets are within 40 nm, the oligonucleotides can ligate and be amplified, generating a fluorescent spot detectable by microscopy. Primary Application: Visualizing and quantifying endogenous, subcellular localization-specific protein-protein interactions with single-molecule sensitivity in fixed cells/tissues.
Detailed Experimental Protocols
Co-IP Protocol for GRP78 Complex Isolation
- Cell Lysis: Harvest HEK293 or relevant cell line (basal or thapsigargin-treated for ER stress). Lyse in 1 mL non-denaturing IP lysis buffer (25mM Tris HCl pH 7.4, 150mM NaCl, 1% NP-40, 1mM EDTA, 5% glycerol) with protease/phosphatase inhibitors. Centrifuge at 16,000×g for 15 min at 4°C.
- Pre-clearing: Incubate supernatant with 20 μL protein A/G agarose beads for 1 hour at 4°C. Centrifuge, collect supernatant.
- Immunoprecipitation: Incubate 500 μg lysate with 2-5 μg anti-GRP78 monoclonal antibody (e.g., clone C50B12) or species-matched IgG control overnight at 4°C with rotation.
- Bead Capture: Add 30 μL protein A/G beads, incubate 2-4 hours at 4°C.
- Washing: Pellet beads, wash 4x with 500 μL ice-cold lysis buffer.
- Elution: Elute bound proteins with 40 μL 2X Laemmli buffer by boiling for 10 min.
- Analysis: Resolve by SDS-PAGE. Perform western blot for candidate interactors (e.g., ATF6, PERK, IRE1α, client proteins) or submit for LC-MS/MS analysis.
Acceptor Photobleaching FRET Protocol
- Sample Prep: Transfect cells with GRP78-CFP (donor) and putative client-YFP (acceptor). Seed on glass-bottom dishes. Optionally, treat with 300 nM thapsigargin for 6h.
- Imaging: Use a confocal microscope with 458 nm (CFP) and 514 nm (YFP) laser lines. Define a region of interest (ROI).
- Pre-bleach Measurement: Capture donor (CFP) and acceptor (YFP) images.
- Acceptor Photobleaching: Bleach YFP in the ROI using high-intensity 514 nm laser.
- Post-bleach Measurement: Immediately capture donor (CFP) image again.
- Analysis: Calculate FRET efficiency (E) per ROI:
E = (D_post - D_pre) / D_post * 100%, where D is donor intensity. An increase in donor fluorescence after acceptor bleaching confirms interaction.
Duolink PLA Protocol for Fixed Cells
- Cell Fixation & Permeabilization: Culture cells on coverslips, treat as required. Fix with 4% PFA for 15 min, permeabilize with 0.1% Triton X-100 for 10 min.
- Blocking: Incubate with Duolink Blocking Solution in a pre-heated humidified chamber for 60 min at 37°C.
- Primary Antibodies: Incubate with a mix of two primary antibodies raised in different species (e.g., mouse anti-GRP78 and rabbit anti-ATF6) diluted in Antibody Diluent overnight at 4°C.
- PLA Probe Incubation: Wash, then add species-specific PLA probes (MINUS and PLUS) for 1 hour at 37°C.
- Ligation & Amplification: Wash, add Ligation-Ligase solution for 30 min at 37°C. Wash, add Amplification-Polymerase solution for 100 min at 37°C.
- Mounting & Imaging: Wash, mount with Duolink In Situ Mounting Medium with DAPI. Acquire z-stacks using a fluorescence microscope. Quantify PLA signals (dots/cell) using ImageJ.
Data Presentation: Quantitative Comparisons
Table 1: Comparison of Key PPI Methodologies for GRP78 Network Mapping
| Parameter | Co-Immunoprecipitation (Co-IP) | FRET (Acceptor Photobleaching) | Proximity Ligation Assay (PLA) |
|---|---|---|---|
| Interaction Proximity | Co-complex membership (~10 nm - >) | Direct interaction (1-10 nm) | Proximal localization (<40 nm) |
| Throughput | Medium-High (MS scale) | Low (pairwise validation) | Medium (multiplexable) |
| Quantification | Semi-quantitative (WB) / Quantitative (MS) | Quantitative (% Efficiency) | Quantitative (dots/cell) |
| Context | Lysates (native or crosslinked) | Live cells | Fixed cells/tissues |
| Key Advantage | Unbiased discovery | Dynamic, direct interaction proof | Endogenous, spatial resolution |
| Typical GRP78 Interactor Validated | PERK, IRE1α, Protein Disulfide Isomerase | Mutant thyroglobulin (client) | ATF6 (in ER stress) |
Table 2: Example FRET Efficiency Data for GRP78-Client Pairs Under ER Stress
| Donor-Acceptor Pair | Basal FRET Efficiency (%) | Thapsigargin-Treated FRET Efficiency (%) | p-value (t-test) | Interpretation |
|---|---|---|---|---|
| GRP78-CFP / ATF6-YFP | 8.2 ± 1.5 | 22.7 ± 3.1 | <0.001 | Stress-dependent interaction increase |
| GRP78-CFP / BSA-YFP (Control) | 1.5 ± 0.8 | 1.8 ± 1.0 | 0.45 | No specific interaction |
| GRP78-CFP / Mutant Proinsulin-YFP | 15.4 ± 2.2 | 31.6 ± 4.5 | <0.001 | Enhanced client chaperone binding |
Visualizations: Pathways and Workflows
Diagram 1: GRP78 Network Mapping Strategy in ER Stress Context
Diagram 2: Acceptor Photobleaching FRET Workflow
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for GRP78 PPI Studies
| Reagent | Function/Application | Example Product (Supplier) |
|---|---|---|
| Anti-GRP78/BiP Antibody (IP-grade) | High-affinity, specific antibody for immunoprecipitation or as a primary in PLA. | Mouse mAb (C50B12) (Cell Signaling #3177) |
| Protein A/G Magnetic Beads | Efficient capture of antibody-protein complexes for Co-IP, reducing background. | Pierce Protein A/G Magnetic Beads (Thermo Fisher) |
| Non-Denaturing Lysis Buffer | Maintains native protein-protein interactions during cell lysis for Co-IP. | IP Lysis Buffer (Thermo Fisher #87788) |
| ER Stress Inducers | Induce GRP78 upregulation and alter its interactome (positive control). | Thapsigargin (Sigma-Aldrich T9033), Tunicamycin (Sigma-Aldrich T7765) |
| Fluorescent Protein Vectors | Cloning donors (CFP) and acceptors (YFP/mCherry) for FRET constructs. | pECFP-C1 & pEYFP-C1 vectors (Takara Bio) |
| Duolink PLA Kit | Complete reagent set for proximity ligation assays on fixed samples. | Duolink In Situ Red Starter Kit (Sigma-Aldrich DUO92101) |
| Protease/Phosphatase Inhibitor Cocktail | Preserves protein integrity and phosphorylation states during lysis. | Halt Cocktail (Thermo Fisher #78440) |
| Fluorescence-Compatible Mounting Medium | Preserves fluorescence signals for FRET/PLA microscopy, often with DAPI. | ProLong Gold Antifade Mountant with DAPI (Thermo Fisher P36931) |
The unfolded protein response (UPR) sensor ATF6 and its primary transcriptional target, the chaperone GRP78/BiP, constitute a critical regulatory axis for endoplasmic reticulum (ER) proteostasis. Within the broader thesis of protein folding research, the ATF6-GRP78 system is not merely a stress reporter but a dynamic, tunable pathway whose activity is intricately linked to disease pathogenesis. In neurodegenerative diseases, chronic ER stress and inadequate UPR signaling contribute to protein aggregation. In cancer, tumors co-opt the pathway to promote survival, proliferation, and therapy resistance. In metabolic disorders, adipocyte and hepatocyte dysfunction is driven by ER stress. Therefore, precise quantification of ATF6-GRP78 activity—through transcriptional, translational, and localization readouts—provides a powerful multidimensional toolkit for modeling disease mechanisms and screening therapeutic interventions.
Quantitative Disease-Associated Alterations in the ATF6-GRP78 Axis
The following table summarizes key quantitative findings from recent studies (2023-2024) highlighting dysregulation of the ATF6-GRP78 axis across disease models.
Table 1: Quantitative Readouts of ATF6-GRP78 Activity in Disease Models
| Disease Model | System/Cell Type | Key Readout | Measurement & Change vs. Control | Implications for Pathogenesis |
|---|---|---|---|---|
| Alzheimer's Disease | APP/PS1 transgenic mouse cortex | GRP78 Protein Level | ↓ 40-50% (Western blot) | Impaired ER folding capacity exacerbates Aβ and tau aggregation. |
| Parkinson's Disease | α-synuclein (A53T) SH-SY5Y cells | Nuclear ATF6 (Active Form) | ↓ 60% (immunofluorescence) | Defective ATF6 activation reduces compensatory chaperone response. |
| Glioblastoma | Patient-derived GBM stem cells | GRP78 Promoter Activity | ↑ 3.5-fold (luciferase reporter) | High basal UPR supports tumor stemness and growth in hypoxic core. |
| Breast Cancer (TNBC) | MDA-MB-231 tumors in mice | Soluble ATF6 (p50) Protein | ↑ 2.8-fold (ELISA of tumor lysate) | Correlates with resistance to doxorubicin chemotherapy. |
| Type 2 Diabetes | High-fat diet mouse liver | ATF6 Target Gene Score | ↑ 4.2-fold (RNA-seq of XBP1s, HERPUD1, etc.) | Chronic nutrient overload induces sustained, maladaptive UPR. |
| NAFLD/NASH | Human steatotic hepatocytes | Cell Surface GRP78 | ↑ 8-fold (FACS analysis) | Drives pro-inflammatory signaling and insulin resistance. |
Core Experimental Protocols for Integrated Readouts
Protocol 1: Simultaneous Measurement of ATF6 Activation and GRP78 Expression (Immunofluorescence & High-Content Imaging)
Objective: To spatially resolve ATF6 nuclear translocation (activation) and concurrent GRP78 upregulation in single cells. Materials: Cells plated on 96-well imaging plates, ER stress inducer (e.g., Tunicamycin 2μg/mL, 6h), control medium. Steps: 1. Fixation & Permeabilization: Aspirate medium, fix with 4% PFA for 15 min, permeabilize with 0.1% Triton X-100 for 10 min. 2. Blocking: Block with 5% BSA in PBS for 1h. 3. Primary Antibody Incubation: Incubate with chicken anti-GRP78 (1:500) and rabbit anti-ATF6 (N-terminal, 1:250) in blocking buffer overnight at 4°C. 4. Secondary Antibody Incubation: Incubate with Alexa Fluor 488-conjugated anti-chicken and Alexa Fluor 594-conjugated anti-rabbit antibodies (1:1000) for 1h. Include DAPI (1μg/mL). 5. Image Acquisition: Acquire images on a high-content imager using 20x objective. Capture 10 fields/well. 6. Analysis: Use image analysis software to: (a) Identify nuclei (DAPI). (b) Measure mean ATF6 fluorescence intensity in the nucleus (AF594). (c) Measure mean GRP78 fluorescence intensity in the cytoplasm (AF488). (d) Calculate the nuclear/cytoplasmic ratio of ATF6.
Protocol 2: GRP78 Promoter-Driven Luciferase Reporter Assay for Pathway Activity
Objective: To quantify the transcriptional output of the ATF6 pathway via its activity on the GRP78 promoter. Materials: Cells, GRP78-promoter-firefly luciferase reporter plasmid (e.g., pGRP78-Luc), Renilla luciferase control plasmid (pRL-TK), transfection reagent, Dual-Glo Luciferase Assay System. Steps: 1. Co-transfection: Seed cells in 96-well plates. Co-transfect with pGRP78-Luc (50ng/well) and pRL-TK (5ng/well) using appropriate transfection reagent. 2. Treatment: 24h post-transfection, treat cells with experimental compounds or stressors for 8-16h. 3. Luciferase Measurement: Aspirate medium, add Dual-Glo Luciferase Reagent, incubate 10 min, measure firefly luminescence (reporter). Then add Dual-Glo Stop & Glo Reagent, incubate 10 min, measure Renilla luminescence (transfection control). 4. Calculation: Normalize firefly luminescence to Renilla luminescence for each well. Express data as fold change relative to untreated control.
Protocol 3: Cell Surface GRP78 Detection by Flow Cytometry
Objective: To quantify the translocation of GRP78 to the cell membrane, a phenomenon linked to oncogenic signaling and metabolic inflammation. Materials: Live cells in suspension, ice-cold FACS buffer (PBS + 2% FBS), anti-GRP78 antibody conjugated to APC (clone specific for extracellular epitope), isotype control-APC. Steps: 1. Harvesting: Harvest cells using gentle, non-enzymatic dissociation to preserve surface proteins. Wash with ice-cold FACS buffer. 2. Staining: Aliquot 1x10^6 cells per tube. Stain with anti-GRP78-APC or isotype control (1:100 dilution) in 100μL FACS buffer for 30 min on ice in the dark. 3. Washing: Wash cells twice with 2mL ice-cold FACS buffer, pellet at 300g for 5 min at 4°C. 4. Analysis: Resuspend in 300μL FACS buffer. Analyze on a flow cytometer using the APC channel. Use isotype control to set the negative gate. Report median fluorescence intensity (MFI) or percentage of GRP78-high cells.
Visualizing the ATF6-GRP78 Pathway and Experimental Integration
Title: ATF6 Activation Pathway Upon ER Stress
Title: GRP78 Promoter Reporter Assay Workflow
The Scientist's Toolkit: Essential Research Reagent Solutions
Table 2: Key Reagents for ATF6-GRP78 Research
| Reagent Category | Specific Item/Example | Function & Application |
|---|---|---|
| Inducers/Inhibitors | Tunicamycin, Thapsigargin, Brefeldin A | Standard pharmacologic ER stressors used to activate the UPR pathways in a controlled manner. |
| ATF6 Modulators | AA147 (Activator), Ceapins (Inhibitor) | Small molecule tools to selectively potentiate or inhibit the ATF6 arm of the UPR for mechanistic studies. |
| Antibodies | Anti-ATF6 (N-terminal, for full-length), Anti-ATF6 (p50, for cleaved), Anti-GRP78/BiP (for IHC/WB/IF), Anti-GRP78 (extracellular epitope for FACS) | Critical for detecting protein expression, localization (nuclear vs. ER), activation state, and surface presentation. |
| Reporter Systems | GRP78 Promoter-Luciferase Plasmid, ERSE (ER Stress Response Element) Reporter | Read transcriptional activity of the pathway. Stable cell lines with these reporters enable HTS. |
| siRNA/shRNA | ATF6-specific, GRP78-specific | For genetic knockdown to validate functional roles of specific components in disease phenotypes. |
| Detection Kits | Dual-Luciferase Reporter Assay Kit, Colorimetric/Flourogenic Protease Activity Kits (for S1P/S2P) | Enable quantitative, normalized measurement of reporter activity or enzymatic steps in ATF6 processing. |
| Cell Lines | ATF6-KO HEK293, GRP78-Haploinsufficient MEFs, Disease-relevant primary cells (e.g., hepatocytes, neurons) | Genetically engineered models to study pathway necessity and physiological context. |
Navigating Experimental Challenges: Optimizing ATF6-GRP78 Pathway Analysis
This technical guide examines the critical challenges in studying the unfolded protein response (UPR), specifically within the context of ATF6-GRP78 chaperone system research. The accurate induction of endoplasmic reticulum (ER) stress is paramount, yet many commonly used chemical stressors exhibit significant off-target effects that confound data interpretation. Furthermore, the complex cross-talk between the IRE1 and PERK branches of the UPR creates a network of compensatory and antagonistic signals that can obscure the specific role of the ATF6 pathway. This document provides an analytical framework to identify, mitigate, and account for these pitfalls in experimental design and data analysis.
Off-Target Effects of Common ER Stress Inducers
ER stress inducers are essential tools, but their mechanisms are often non-specific.
Quantitative Analysis of Common Stressors
The table below summarizes the primary and documented off-target effects of standard pharmacological ER stressors.
Table 1: Profiles of Common Pharmacological ER Stressors
| Stressor (Concentration Range) | Primary Target | Key Off-Target Effects | Impact on ATF6-GRP78 System |
|---|---|---|---|
| Tunicamycin (0.1-10 µg/mL) | N-linked glycosylation (inhibits GlcNAc phosphotransferase) | Activates DNA damage response; Alters cell adhesion; Induces oxidative stress. | Potent and relatively specific ATF6 activation due to pure protein misfolding. GRP78 dissociation is direct. |
| Thapsigargin (10-300 nM) | SERCA pump (Sarco/Endoplasmic Reticulum Ca²⁺ ATPase) | Disrupts mitochondrial Ca²⁺ homeostasis; Alters cytosolic signaling (NF-κB, etc.); Induces apoptosis via non-UPR pathways. | Strong ATF6 activation via Ca²⁺ depletion and GRP78 dissociation. PERK/IRE1 also strongly activated, leading to cross-talk. |
| Brefeldin A (0.1-10 µM) | ARF-GEF (inhibits ER-to-Golgi transport) | Disrupts trans-Golgi network; Induces lysosomal stress; Alters lipid metabolism. | Weak/moderate ATF6 activation. Major confounder is blockade of ATF6 transport to Golgi, inhibiting its cleavage. |
| Dithiothreitol (DTT) (1-5 mM) | Reduces disulfide bonds | General redox disruptor; Affects cytoplasmic and mitochondrial proteins; Induces necrosis at high doses. | Rapid ATF6 activation due to misfolding of disulfide-bonded proteins. Effects are acute and can be toxic. |
Experimental Protocol: Validating Stressor Specificity
To control for off-target effects, a combinatorial validation approach is recommended.
Protocol: Specificity Validation for ER Stress Induction
- Cell Treatment: Treat your model system (e.g., HEK293, HeLa, primary hepatocytes) with the chosen stressor at its standard EC₅₀ for UPR induction (e.g., 2 µg/mL Tunicamycin, 100 nM Thapsigargin) for 6 hours.
- Multi-Parameter Assessment:
- UPR Marker Analysis: By western blot, probe for canonical markers of all three branches: ATF6 (cleaved p50 fragment), IRE1 (XBP1 splicing via RT-PCR or anti-XBP1s antibody), and PERK (phospho-eIF2α, ATF4).
- Off-Target Marker Analysis: In parallel, probe for markers of confounding pathways: DNA damage (γH2AX), oxidative stress (Nrf2, HO-1), and general apoptosis (cleaved Caspase-3).
- Genetic Confirmation: Use siRNA knockdown of key UPR sensors (IRE1, PERK, ATF6). A specific inducer's effect on a downstream marker (e.g., CHOP for PERK) should be attenuated only by knockdown of its primary sensor.
- GRP78 Dissociation Assay: Perform co-immunoprecipitation in untreated and stressed cells using an anti-ATF6 antibody. A true ER stressor should reduce the amount of GRP78 co-precipitated with ATF6, indicating their dissociation.
Cross-Talk Between IRE1, PERK, and ATF6 Pathways
The UPR branches do not operate in isolation. Signaling convergence and feedback loops create a complex network.
Key Nodes of Pathway Interaction
Table 2: Documented Cross-Talk Mechanisms Between UPR Branches
| Cross-Talk Interaction | Molecular Mechanism | Consequence for ATF6-GRP78 Studies |
|---|---|---|
| IRE1 attenuates PERK signaling | IRE1-mediated decay (RIDD) of Perk and Atf4 mRNAs. | Under prolonged stress, IRE1 activity can dampen the PERK response, potentially shifting cellular fate. May alter the balance between adaptive (ATF6) and pro-apoptotic (CHOP) outputs. |
| PERK inhibits IRE1 signaling | eIF2α phosphorylation reduces global translation, including IRE1 and XBP1 synthesis. | Early PERK activation can limit the capacity of the IRE1-XBP1 axis, another adaptive arm. This can make cells more dependent on ATF6 for chaperone production. |
| Shared Downstream Targets | XBP1s (IRE1) and ATF6(p50) both bind to ERSE/UPRE promoters to regulate overlapping gene sets (e.g., GRP78, PDI). | Upregulation of GRP78 cannot be solely attributed to ATF6 activation. Genetic or chemical inhibition of IRE1 is required to isolate the ATF6-specific contribution. |
| Feedback Inhibition by GRP78 | Resynthesized GRP78 binds to and inactivates all three sensors. | This is a universal negative feedback loop. Measuring GRP78 protein levels and its re-association with ATF6 is crucial for understanding the timing and attenuation of the response. |
Diagram 1: UPR Branch Cross-Talk and GRP78 Feedback.
Experimental Protocol: Disentangling Cross-Talk
To isolate the ATF6-specific response, targeted inhibition of IRE1 and PERK is necessary.
Protocol: Isolating the ATF6 Transcriptional Response
- Pharmacological Inhibition:
- IRE1 Inhibition: Pre-treat cells with 10-50 µM 4µ8C (IRE1 RNase inhibitor) or 1-10 µM STF-083010 for 1 hour before ER stress induction.
- PERK Inhibition: Pre-treat cells with 1 µM GSK2606414 (PERK inhibitor) or use a PERK-knockout cell line.
- Induction & Analysis: Induce ER stress with a chosen agent (e.g., Thapsigargin). Harvest cells at multiple time points (e.g., 2, 6, 12h).
- Readout: Perform qRT-PCR on a panel of UPR target genes. Classify them as:
- ATF6-Primary: Genes whose induction is unchanged by IRE1/PERK inhibitors but is abolished by ATF6 siRNA (e.g., SARP1).
- XBP1-Primary: Genes whose induction is blocked by IRE1 inhibitor (e.g., EDEM1).
- Composite: Genes whose induction is partially reduced by both IRE1 and ATF6 inhibition (e.g., GRP78). The residual signal with IRE1 inhibited is the ATF6 contribution.
Diagram 2: Workflow to Isolate ATF6-Specific Gene Expression.
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for UPR and ATF6-GRP78 Research
| Reagent / Material | Function & Application | Key Considerations |
|---|---|---|
| Tunicamycin (≥98% HPLC) | Gold-standard for inducing ER stress via N-glycosylation blockade. Used to study pure protein misfolding response. | Verify purity; prepare fresh stock in DMSO or NaOH; high cytotoxicity. |
| Thapsigargin (High Purity) | Potent SERCA pump inhibitor causing ER Ca²⁺ depletion. Used for strong, rapid UPR induction across all branches. | Potent biohazard; use nanomolar concentrations; significant off-target Ca²⁺ effects. |
| 4µ8C | Selective, cell-permeable inhibitor of IRE1's RNase activity. Critical for blocking the IRE1-XBP1/RIDD axis in cross-talk studies. | Does not affect IRE1 kinase activity; confirm inhibition via XBP1 splicing assay. |
| GSK2606414 or GSK2656157 | Potent and selective ATP-competitive inhibitors of PERK kinase activity. Essential for isolating non-PERK UPR signaling. | Monitor for compensatory IRE1 hyperactivation; use appropriate vehicle controls. |
| Anti-ATF6 (Cleaved p50) Antibody | Detects the active, nuclear form of ATF6 by western blot. Primary readout for ATF6 pathway activation. | Many antibodies detect full-length only; validation with a positive control (e.g., Tm/Tg treatment) is mandatory. |
| GRP78/BiP Co-IP Kit | Immunoprecipitation reagents to study the interaction between GRP78 and ATF6/IRE1/PERK under stress vs. resting conditions. | Demonstrates direct mechanistic link between misfolded protein load, chaperone dissociation, and sensor activation. |
| ATF6α Knockout Cell Line | Genetic model (CRISPR/Cas9) to conclusively determine ATF6-dependent phenotypes and gene regulation. | Required for definitive attribution; ensure off-target effects of CRISPR are controlled. |
| ER-Targeted GFP (ssGFP-HDEL) | Reporter construct to visualize ER morphology and dilation in real-time upon ER stress induction. | Useful for correlating functional stress with biochemical UPR markers. |
Within the broader thesis on the ATF6-GRP78 chaperone system in protein folding research, a critical methodological challenge is the precise dissection of ATF6-specific signaling branches from the integrated unfolded protein response (UPR). The UPR, initiated by ER stress, is transduced by three principal sensors: IRE1α, PERK, and ATF6. While these pathways coordinate to restore proteostasis, their individual contributions—particularly those of ATF6—to transcriptional programs, chaperone induction, and cell fate decisions are often conflated. This guide provides a technical framework for isolating and validating ATF6 activation, separating it from parallel UPR arms and off-target effects.
The UPR Signaling Network: Overlap and Distinction
The ATF6 pathway is uniquely defined by its regulated intramembrane proteolysis. Under ER stress, dissociation of the chaperone GRP78 from ATF6 allows its translocation to the Golgi apparatus, where it is cleaved by Site-1 Protease (S1P) and Site-2 Protease (S2P). The liberated ATF6(N) cytosolic domain translocates to the nucleus to activate genes harboring ER stress response elements (ERSE). Key target genes include GRP78/BiP, XBP1, and chaperones like PDI and GRP94. This pathway overlaps with IRE1α-XBP1s and PERK-ATF4 signaling, necessitating specific interrogation tools.
Diagram 1: Integrated UPR with ATF6 Cleavage
Key Quantitative Markers for Pathway Specificity
The table below summarizes core quantitative outputs used to distinguish ATF6 activation from general UPR. Data is compiled from recent literature (2022-2024).
Table 1: Discriminatory Markers of UPR Sensor Activation
| Parameter | ATF6-Specific | IRE1α-XBP1s | PERK-ATF4 | Measurement Technique |
|---|---|---|---|---|
| Key Transcriptional Target | GRP78 (early), PDIA4 | XBP1s (spliced), EDEM1 | CHOP, ATF4, GADD34 | qRT-PCR, RNA-seq |
| Proteolytic Fragment | ATF6(N) p50 (~50 kDa) | XBP1s (54 kDa) | ATF4 (38 kDa) | Western Blot (Cleavage-specific Abs) |
| Canonical Signaling Element | ERSE Promoter Element | UPRE Promoter Element | AARE Promoter Element | Luciferase Reporter Assay |
| Kinetics of Activation | Intermediate (1-4h peak) | Fast (30min-2h peak) | Sustained (2-8h peak) | Time-course immunoblot |
| Pharmacologic Inhibitor | Ceapins (block S1P cleavage) | 4μ8C (IRE1 RNase inhibitor) | GSK2606414 (PERK inhibitor), ISRIB (blocks eIF2α-P effects) | Inhibitor + Stressor co-treatment |
| Genetic Knockout Phenotype (Mouse) | Impaired chaperone induction, steatosis | Impaired plasma cell differentiation | Pancreatic dysfunction, neurodegeneration | Phenotypic analysis post-ER stress |
Experimental Protocols for Isolating ATF6 Signaling
Protocol: Detection of ATF6 Cleavage by Immunoblotting
Objective: To specifically monitor the proteolytic activation of ATF6. Materials: Anti-ATF6α antibody (clone 1G7, C-terminal), anti-β-actin antibody, RIPA buffer, proteasome inhibitor (MG132), S1P inhibitor (PF-429242 or Ceapin-A7). Procedure:
- Treat cells (e.g., HEK293, HepG2) with ER stress inducers: Tunicamycin (Tm, 2 µg/mL, inhibits N-glycosylation) or Thapsigargin (Tg, 300 nM, SERCA inhibitor). Include inhibitor controls (e.g., Ceapin-A7, 10 µM, added 30 min prior).
- At time points (0, 1, 2, 4, 8h), lyse cells in RIPA buffer supplemented with protease inhibitors.
- Centrifuge lysates (12,000 x g, 15 min, 4°C).
- Resolve 30 µg protein on 4-12% Bis-Tris gels and transfer to PVDF membrane.
- Block with 5% BSA, then probe with anti-ATF6α antibody (1:1000). The full-length p90 and cleaved cytosolic p50 fragments are detected.
- Specificity Control: Co-treatment with Ceapin-A7 should block the appearance of the p50 band without affecting Tm/Tg-induced p90 upregulation.
Protocol: ATF6-Specific Transcriptional Reporter Assay
Objective: To quantify ATF6-driven transcriptional activity distinct from IRE1 or PERK. Materials: pGL4-ERSE-Luciferase reporter plasmid (3x ERSE consensus sequence), pRL-TK Renilla control, Dual-Luciferase Reporter Assay System. Procedure:
- Seed cells in 24-well plates. Co-transfect with pGL4-ERSE-Luciferase (firefly) and pRL-TK (Renilla) plasmids using standard transfection reagent.
- After 24h, treat cells with ER stressors and/or pathway inhibitors (see Table 1).
- After 8-16h, harvest cells and perform dual-luciferase assay per manufacturer's protocol.
- Specificity Control: Normalize Firefly luminescence to Renilla. Activity should be abrogated by ATF6 siRNA co-transfection or S1P inhibitor, but remain largely unaffected by IRE1/PERK-specific inhibitors.
Diagram 2: ATF6-Specific Reporter Assay Workflow
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Specific ATF6 Research
| Reagent | Category | Function in ATF6 Research | Example (Supplier) |
|---|---|---|---|
| Ceapin-A7 | Small Molecule Inhibitor | Specifically blocks S1P-mediated cleavage of ATF6 in the Golgi, without affecting SREBP processing or other UPR arms. | Tocris (Cat. No. 5770) |
| Tunicamycin (Tm) | ER Stress Inducer | Inhibits N-linked glycosylation, causing ER protein misfolding and robust activation of all UPR sensors, including ATF6. | Sigma-Aldrich (Cat. No. T7765) |
| Anti-ATF6α (1G7) | Antibody | Monoclonal antibody recognizing C-terminus of ATF6α; detects both full-length (p90) and cleaved (p50) forms by Western blot. | Novus Biologicals (Cat. No. NBP1-40256) |
| pGL4-ERSE-Luc | Reporter Plasmid | Plasmid containing multiple ERSE elements driving Firefly luciferase; primary tool for quantifying ATF6-specific transcriptional output. | Addgene (Plasmid #11976) |
| PF-429242 | S1P Inhibitor | Inhibits Site-1 Protease, blocking cleavage of both ATF6 and SREBP; used as a less-specific but established ATF6 pathway blocker. | MedChemExpress (Cat. No. HY-13423) |
| ATF6α siRNA Pool | Genetic Tool | Specific knockdown of ATF6α expression to confirm the dependency of observed effects on this isoform. | Dharmacon (ON-TARGETplus) |
| Recombinant GRP78/BiP | Protein | Used in binding/competition assays to study the dynamics of GRP78-ATF6 dissociation. | Abcam (Cat. No. ab78432) |
Data Integration and Concluding Remarks
Robust distinction of ATF6 signaling requires a multi-parametric approach: 1) monitoring the p50 cleavage fragment, 2) using ERSE-specific reporters with pharmacological/genetic inhibition, and 3) validating with a subset of ATF6 target genes (e.g., PDIA4). Researchers must account for cell-type-specific UPR dynamics and the partial functional redundancy between XBP1s and ATF6(N). The protocols and tools outlined here, when applied within the context of the ATF6-GRP78 chaperone axis, enable precise mechanistic dissection critical for developing therapies targeting specific UPR branches in protein misfolding diseases.
1. Introduction The study of the ATF6-GRP78 chaperone system, a critical branch of the unfolded protein response (UPR), is fundamental to understanding protein folding homeostasis in health and disease. Precise detection of these key proteins (ATF6, its cleaved active form, and GRP78/BiP) via Western blot (WB) and immunofluorescence (IF) is paramount. However, challenges like antibody cross-reactivity, high background, and poor signal-to-noise (S/N) ratios often obscure results. This guide provides a technical framework for optimizing detection specificity and sensitivity, framed within ATF6-GRP78 research.
2. Core Challenges in ATF6/GRP78 Detection
- ATF6: Distinguishing full-length (p90ATF6α, ~90 kDa) from the cleaved, active nuclear form (p50ATF6α, ~50 kDa) is essential but challenging due to homologous proteins and cleavage fragments.
- GRP78: This abundant chaperone exists in multiple states (free, substrate-bound). Detection shifts can be subtle, requiring high specificity.
- Common Issues: Non-specific binding to other ER chaperones (e.g., GRP94, calnexin) or UPR-related transcription factors plagues both WB and IF.
3. Quantitative Data Summary: Impact of Optimization Steps Table 1: Quantitative Impact of Optimization Techniques on Signal-to-Noise Ratio
| Optimization Technique | Application | Typical S/N Improvement* | Key Metric |
|---|---|---|---|
| Antody Validation (KO validation) | WB, IF | 5-10 fold | Specificity Index (Wild-type/KO signal) |
| Blocking Buffer Optimization | WB, IF | 2-5 fold | Background Optical Density (OD) |
| Stringency Wash Optimization | IF | 3-8 fold | Fluorescence Intensity (Specific/Non-specific) |
| Tyramide Signal Amplification | IF | 10-100 fold | Detection Limit (Lowest antigen copy #) |
| High-Dynamic Range Imaging | WB | 2-4 fold | Linear Dynamic Range (Orders of magnitude) |
*S/N improvement is dependent on starting conditions and sample type.
4. Experimental Protocols for Key Optimization Steps
Protocol 4.1: Knockout-Validated Antibody Screening for ATF6/GRP78 Objective: Confirm antibody specificity using ATF6 or HSPA5 (GRP78) knockout cell lines. Materials: Wild-type (WT) and KO cell lysates (e.g., HEK293 ATF6α/β DKO), candidate antibodies, standard WB reagents. Method:
- Prepare lysates from WT and KO cells treated with/without ER stress inducer (e.g., 2µg/mL Tunicamycin, 16h).
- Run 30-50 µg total protein per lane on a 4-12% Bis-Tris gel.
- Transfer to PVDF membrane.
- Block with 5% BSA/TBST for 1h.
- Incubate with primary antibody (diluted in block) overnight at 4°C.
- Wash 3x with TBST, incubate with HRP-conjugated secondary (1h, RT).
- Develop with high-sensitivity chemiluminescent substrate. Validation: A valid antibody shows a strong signal in WT lanes (correct molecular weight) and absence of signal in KO lanes at the same position.
Protocol 4.2: Enhanced Stringency Immunofluorescence for GRP78 Localization Objective: Reduce background for clear ER-specific GRP78 staining. Materials: Fixed cells, validated anti-GRP78 antibody, fluorescence secondary, PBS, Triton X-100. Method:
- Culture and treat cells on chambered coverslips. Fix with 4% PFA (15 min, RT).
- Permeabilize with 0.1% Triton X-100/PBS (10 min).
- Block: Use 5% normal serum from secondary host + 1% BSA in PBS (1h).
- Primary Antibody: Incubate in block at optimized concentration (2h, RT).
- Stringency Washes: Wash 3x with PBS. Perform a critical wash with PBS containing 0.05% Tween-20 and 0.1% Triton X-100 for 10 min with gentle agitation.
- Incubate with fluorophore-conjugated secondary (1h, RT, in dark).
- Wash 3x with PBS, include DAPI, mount. Key: The critical wash (Step 5) disrupts weak, non-specific ionic/hydrophobic interactions without eluting high-affinity specific bonds.
5. Visualization of Pathways and Workflows
Diagram 1: ATF6 Pathway Activation Upon ER Stress
Diagram 2: Antibody Validation Workflow
6. The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Optimized ATF6/GRP78 Detection
| Reagent / Solution | Primary Function | Application | Example & Rationale |
|---|---|---|---|
| KO-Validated Primary Antibodies | Binds target antigen with high specificity; confirmed using genetic controls. | WB, IF | Commercial antibodies validated in ATF6α/β DKO or HSPA5 KO cells. Non-validated antibodies are high-risk. |
| High-Sensitivity Chemiluminescent Substrate | Amplifies HRP enzyme signal for low-abundance target detection. | WB | ECL Prime or SuperSignal West Femto. Essential for detecting cleaved p50ATF6α. |
| Tyramide Signal Amplification (TSA) Kits | Enzyme-mediated deposition of fluorophores for ultra-sensitive target detection. | IF (Multiplex) | Opal TSA. Can detect subtle GRP78 expression shifts. |
| Tissue Culture-Grade ER Stress Inducers | Induces the UPR pathway specifically and reproducibly. | Sample Prep | Tunicamycin (N-glycosylation block), Thapsigargin (SERCA inhibitor). Positive control for ATF6 cleavage. |
| Normal Serum (from Secondary Host) | Blocks non-specific binding of secondary antibodies to sample proteins. | IF Blocking | Use 5% normal goat serum if secondary is goat-anti-rabbit. Reduces background significantly. |
| High-Stringency Wash Buffer | Removes low-affinity, non-specifically bound antibodies while retaining specific binding. | IF Washes | PBS with 0.05% Tween-20 + 0.1% Triton X-100. Critical step post-primary antibody incubation. |
Within the broader thesis on the ATF6-GRP78 chaperone system in protein folding research, this whitepaper examines the critical influence of cellular context and model systems on the observed activation and downstream signaling of the Unfolded Protein Response (UPR) branch. Empirical data consistently demonstrates that the magnitude, kinetics, and functional outcomes of ATF6-mediated pathways are not uniform but are significantly modulated by cell type, disease state, and experimental model. This guide synthesizes current findings, providing a technical framework for designing, interpreting, and comparing studies on this adaptive signaling network.
The endoplasmic reticulum (ER) chaperone GRP78 (BiP) is a master regulator of ER stress sensors, including ATF6. Under homeostatic conditions, GRP78 binds to ATF6, retaining it in the ER. Accumulation of unfolded proteins sequesters GRP78, permitting ATF6 translocation to the Golgi apparatus where it is cleaved to release its active cytosolic fragment (ATF6f). ATF6f translocates to the nucleus to upregulate chaperone genes, including GRP78 itself, forming a critical feedback loop. This system's core biology is conserved, yet its operational parameters are highly context-dependent.
Quantitative Evidence of Variable Responses
The following tables summarize key comparative data illustrating differential ATF6 pathway responses.
Table 1: ATF6 Activation Kinetics and Magnitude Across Cell Types
| Cell Type / Model | Inducer (Concentration) | Time to Peak ATF6f (hr) | Fold Increase in GRP78 mRNA | Key Contextual Note |
|---|---|---|---|---|
| Primary Mouse Hepatocytes | Tunicamycin (5 µg/mL) | 4-6 | 8.2 ± 1.5 | Robust adaptive response |
| HEK293T (Human Embryonic Kidney) | Thapsigargin (1 µM) | 2-3 | 12.5 ± 2.1 | High basal ER load |
| SH-SY5Y (Human Neuroblastoma) | Tunicamycin (2 µg/mL) | 6-8 | 5.0 ± 0.8 | Sensitive to prolonged stress; apoptosis-prone |
| MCF-7 (Breast Cancer) | DTT (2 mM) | 3-4 | 15.3 ± 3.0 | Hyper-activated UPR; pro-survival bias |
| Primary Rat Cardiomyocytes | Hypoxia (1% O₂) | 8-10 | 3.5 ± 0.7 | Slow, attenuated response |
Table 2: Differential Functional Outcomes of ATF6 Activation
| Disease Model | Genetic/Perturbation Context | ATF6 Pathway Activity | Observed Phenotype | Contrast with Other Models |
|---|---|---|---|---|
| Cancer (Glioblastoma) | High GRP78 expression | Constitutively active | Proliferation, chemo-resistance | In normal astrocytes, activation inhibits proliferation. |
| Neurodegeneration (Alzheimer's model) | APP overexpression | Initially elevated, then suppressed | Early protection, late failure | In non-neuronal cells, sustained activation is achievable. |
| Metabolic Disease (Liver Steatosis) | High-fat diet | Attenuated/Desensitized | ER stress persistence, inflammation | In healthy liver, acute activation resolves stress. |
Detailed Experimental Protocols
Protocol: Monitoring ATF6 Cleavage and Translocation (Immunofluorescence & Western Blot)
Objective: To assess ATF6 activation dynamics in different cell lines. Key Reagents: Thapsigargin (SERCA inhibitor), Tunicamycin (N-glycosylation inhibitor), Anti-ATF6 antibody (full length), Anti-ATF6f antibody (cleaved), Golgi marker (GM130), Nuclear dye (DAPI). Procedure:
- Culture & Stress Induction: Plate cells (e.g., HEK293T vs. SH-SY5Y) on coverslips in 12-well plates. At 70% confluence, treat with vehicle (control) or ER stress inducer (e.g., 1µM Thapsigargin) for varying durations (1, 2, 4, 8h).
- Fixation & Permeabilization: Aspirate medium, wash with PBS, and fix with 4% paraformaldehyde (15 min). Permeabilize with 0.1% Triton X-100 (10 min).
- Immunostaining: Block with 5% BSA (1h). Incubate with primary antibodies: anti-ATF6 (1:500) and anti-GM130 (1:1000) overnight at 4°C. Wash, then incubate with species-appropriate fluorescent secondary antibodies (1:1000) and DAPI (1 µg/mL) for 1h.
- Imaging & Analysis: Image using a confocal microscope. Quantify the ratio of nuclear (ATF6f) to ER/Golgi (full-length ATF6) fluorescence intensity per cell using ImageJ software.
- Western Blot Validation: In parallel, harvest cell lysates. Separate proteins via SDS-PAGE (10% gel). Transfer to PVDF membrane and probe sequentially with anti-ATF6f (to detect cleavage) and β-actin loading control.
Protocol: Quantitative Assessment of Transcriptional Output (RT-qPCR)
Objective: To measure cell-type-specific transcriptional responses downstream of ATF6. Procedure:
- RNA Extraction: After stress induction, lyse cells in TRIzol. Isolate total RNA following manufacturer's protocol. Determine purity and concentration via Nanodrop.
- cDNA Synthesis: Use 1 µg of total RNA with a reverse transcription kit including oligo(dT) primers.
- qPCR Setup: Prepare reactions with SYBR Green master mix, gene-specific primers (e.g., GRP78, CHOP, XBP1s), and cDNA template. Use GAPDH or ACTB as housekeeping controls.
- Data Analysis: Calculate fold change using the 2^(-ΔΔCt) method. Compare the kinetics and amplitude of GRP78 induction across cell types.
Visualization of Pathways and Workflows
Title: ATF6 Activation Pathway from ER Stress to Gene Transcription
Title: Workflow for Comparing ATF6 Responses Across Models
The Scientist's Toolkit: Key Research Reagent Solutions
| Reagent / Material | Function in ATF6/GRP78 Research | Key Consideration for Model Variability |
|---|---|---|
| ER Stress Inducers (Tunicamycin, Thapsigargin, DTT) | To perturb ER homeostasis and trigger the UPR. | Cell-type-specific toxicity thresholds must be determined via dose-response curves. |
| ATF6 Activation Inhibitor (Ceapins) | Specifically blocks the S1P protease-mediated cleavage of ATF6. | Useful for isolating ATF6's role from IRE1/XBP1 and PERK branches in complex responses. |
| Anti-ATF6 (Full Length) Antibody | Detects inactive ER-resident ATF6 via WB/IF. | Localization patterns (ER vs. Golgi) are context-dependent; requires careful optimization. |
| Anti-ATF6f (Cleaved) Antibody | Specifically detects the active nuclear form of ATF6. | Critical for assessing activation kinetics, which vary significantly between models. |
| GRP78/BiP Reporter Plasmid (GRP78-promoter-luciferase) | Measures ATF6-mediated transcriptional activity. | Basal promoter activity differs; normalize to a constitutive control. |
| GRP78 siRNA / shRNA | Knocks down GRP78 to disrupt the feedback loop. | Essential for probing adaptive capacity; cells with high basal stress may undergo apoptosis. |
| Site-2 Protease (S2P) Inhibitor | Blocks ATF6 cleavage in the Golgi. | Confirms Golgi-dependent processing; efficacy may depend on Golgi integrity in diseased cells. |
| ATF6 Target Gene qPCR Array | Profiles a panel of ATF6-regulated chaperones and ERAD components. | Identifies cell-type-specific transcriptional programs beyond GRP78. |
This whitepaper examines the critical molecular boundary separating adaptive endoplasmic reticulum (ER) stress signaling from the initiation of apoptosis, framed within the broader thesis on the centrality of the ATF6-GRP78 chaperone system in proteostasis research. The ER responds to the accumulation of unfolded proteins (Unfolded Protein Response, UPR) through sensors like ATF6, IRE1α, and PERK. A key hypothesis in the field posits that the GRP78 chaperone is not merely a passive regulator but the master rheostat that determines the transition from ATF6-mediated adaptive gene expression to pro-apoptotic CHOP activation. Decoupling these pathways quantitatively is essential for therapeutic interventions in diseases ranging from neurodegeneration to cancer.
Core Signaling Pathways and Molecular Thresholds
The ATF6-GRP78 Regulatory Axis
Under homeostatic conditions, GRP78 binds to and inhibits ATF6. ER stress leads to GRP78 sequestration by misfolded proteins, releasing ATF6. ATF6 traffics to the Golgi, is cleaved (ATF6f), and translocates to the nucleus to upregulate chaperones (GRP78, GRP94) and ER-associated degradation (ERAD) components. This defines the adaptive phase.
The Transition to Apoptosis
Sustained, unresolvable stress leads to persistent PERK activation, resulting in prolonged translation attenuation and transcriptional upregulation of the transcription factor CHOP. CHOP drives expression of pro-apoptotic proteins (e.g., BIM, DR5). The decoupling challenge lies in identifying the quantitative tipping point where ATF6-driven adaptation fails and CHOP-driven apoptosis commits.
Diagram 1: ER Stress Decision Pathway: Adaptation vs. Apoptosis
Quantitative Data Interpretation
Key experimental readouts for decoupling involve measuring the dynamics and amplitude of pathway-specific markers.
Table 1: Quantitative Markers for Adaptive vs. Pro-Apoptotic ER Stress
| Signaling Arm | Key Marker | Adaptive Phase (Low/Moderate Stress) | Pro-Apoptotic Phase (Severe/Prolonged Stress) | Measurement Technique |
|---|---|---|---|---|
| ATF6 Axis | GRP78 mRNA | Early, sustained increase (3-5 fold) | Plateaus or declines | qRT-PCR |
| Nuclear ATF6f | Transient peak (1-3 hr post-stress) | Persistent or absent | Western Blot (Nuclear Fraction) | |
| PERK Axis | p-eIF2α | Transient increase (<6 hr) | Sustained elevation (>12 hr) | Phospho-Western Blot |
| CHOP mRNA | Low or modest increase (<10 fold) | High, sustained increase (>25 fold) | qRT-PCR | |
| Integrated Outcome | Caspase-3/7 Activity | Baseline | >5-fold increase over baseline | Luminescent Assay |
| Cell Viability | >85% | <60% | ATP-based Assay |
Table 2: Experimental Tipping Points in Model Cell Lines (Tunicamycin Treatment)
| Cell Line | ATF6-Adaptive Threshold (Tunicamycin nM, duration) | CHOP-Apoptotic Threshold (Tunicamycin nM, duration) | Critical GRP78 Depletion Level | Primary Reference |
|---|---|---|---|---|
| HEK293 | 250 nM, 8h | 500 nM, 16h | ~40% of basal | Shoulders et al., 2013 |
| HeLa | 100 nM, 6h | 300 nM, 12h | ~50% of basal | † |
| INS-1 (Pancreatic β) | 50 nM, 4h | 150 nM, 10h | ~30% of basal | † |
| SH-SY5Y (Neuronal) | 75 nM, 6h | 200 nM, 14h | ~35% of basal | † |
† Compiled from recent literature (2021-2023).
Detailed Experimental Protocols
Protocol: Quantifying the ATF6-CHOP Decision Threshold
Objective: To define the stress dose and duration that decouples adaptive ATF6 signaling from CHOP-driven apoptosis. Reagents: Tunicamycin (ER stressor); Thapsigargin (SERCA inhibitor); ATF6α Antibody (Cell Signaling #65880); CHOP Antibody (CST #5554); GRP78/BiP Antibody (CST #3177).
Procedure:
- Cell Seeding & Stress Induction: Seed HEK293 cells in 12-well plates. At 80% confluency, treat with a Tunicamycin gradient (0, 50, 100, 250, 500, 1000 nM) in triplicate.
- Time-Course Harvest: Harvest cells at 0, 2, 4, 8, 12, 16, and 24h post-treatment for both RNA and protein.
- ATF6 Processing Assay (Western Blot):
- Lyse cells in RIPA buffer with protease inhibitors.
- For nuclear ATF6f, isolate nuclear fractions using a commercial kit (e.g., NE-PER).
- Run 30μg protein on 10% Bis-Tris gel, transfer to PVDF.
- Probe with anti-ATF6α (1:1000) to detect full-length (90 kDa) and cleaved nuclear form (50 kDa).
- Transcriptional Output (qRT-PCR):
- Extract total RNA, synthesize cDNA.
- Perform qPCR using SYBR Green with primers for HSPA5 (GRP78), DDIT3 (CHOP), and ACTB (control).
- Calculate fold change via ΔΔCt method.
- Apoptosis Commitment Assay: At 24h, measure Caspase-3/7 activity using a luminescent substrate (e.g., Caspase-Glo 3/7).
- Data Integration: Plot GRP78/CHOP mRNA ratio vs. Caspase-3/7 activity. The inflection point indicates the decoupling threshold.
Diagram 2: Experimental Workflow: Decoupling Threshold Assay
Protocol: Modulating the GRP78 Rheostat with siRNA
Objective: To directly test GRP78's role in setting the apoptotic threshold. Procedure:
- Transfect cells with HSPA5-targeting siRNA or non-targeting control using lipid nanoparticles.
- 48h post-transfection, confirm GRP78 knockdown (≥70%) by Western blot.
- Subject knockdown and control cells to sub-apoptotic ER stress (e.g., 150 nM Tunicamycin, 8h).
- Measure CHOP induction and Caspase-3/7 activity. Expected Outcome: GRP78 knockdown shifts the threshold, causing a pro-apoptotic response to a normally adaptive stress dose.
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Decoupling ER Stress Research
| Reagent/Material | Supplier Examples | Function in Research | Key Application |
|---|---|---|---|
| ER Stress Inducers | |||
| Tunicamycin | Sigma-Aldrich, Cayman Chemical | N-linked glycosylation inhibitor; induces canonical ER stress. | Titratable stressor for threshold assays. |
| Thapsigargin | Tocris, Abcam | SERCA pump inhibitor; causes ER calcium depletion & stress. | Alternative stress inducer. |
| Molecular Probes & Antibodies | |||
| Anti-ATF6α (Full Length/Cleaved) | Cell Signaling #65880, Abcam ab122897 | Detects both inactive and active nuclear ATF6. | Key for monitoring ATF6 axis activation. |
| Anti-GRP78/BiP | CST #3177, BD Biosciences #610978 | Labels the master chaperone regulator. | Quantifying GRP78 sequestration/induction. |
| Anti-CHOP (DDIT3) | CST #5554, Santa Cruz sc-7351 | Marker for pro-apoptotic UPR output. | Defining apoptotic commitment. |
| Assay Kits | |||
| Caspase-Glo 3/7 Assay | Promega | Luminescent measurement of effector caspase activity. | Quantifying apoptotic commitment. |
| Xbp1 Splicing Assay | (Custom primers/RT-PCR) | Detects IRE1α activation via Xbp1 mRNA splicing. | Monitoring the IRE1α arm. |
| Genetic Tools | |||
| HSPA5 (GRP78) siRNA | Dharmacon, Qiagen | Knocks down GRP78 expression. | Testing GRP78 rheostat function. |
| ATF6 Reporter Plasmid (p5xATF6-GL3) | Addgene #11976 | Luciferase reporter for ATF6 transcriptional activity. | Quantifying ATF6 output independently. |
Precise data interpretation mandates moving beyond qualitative "UPR activation" to a quantitative model of pathway flux. The ATF6-GRP78 axis is not just adaptive; its capacity and dynamics define the system's breaking point. Successful decoupling in disease contexts—such as enhancing adaptive signaling in neurons or forcing apoptotic tipping in cancer cells—requires targeting nodes that specifically widen the gap between these thresholds, with GRP78 itself being the most promising master regulator.
Validating Therapeutic Targets: Comparing ATF6 to Other UPR Pathways in Disease
Within the broader thesis on the ATF6-GRP78 chaperone system in protein folding and endoplasmic reticulum (ER) stress response, the validation of ATF6 activation remains a critical challenge. ATF6, a key sensor of ER stress, translocates to the Golgi upon activation, where it is cleaved to release its cytosolic fragment (ATF6f). This fragment functions as a transcription factor, upregulating chaperones like GRP78/BiP and other genes in the unfolded protein response (UPR). Accurate, multi-faceted validation is essential for research into protein misfolding diseases and for drug development targeting this pathway. This guide details a suite of orthogonal assays to benchmark ATF6 activation, ensuring robust and reproducible conclusions.
Orthogonal Assay Methodologies for ATF6 Activation
Orthogonal validation employs independent methods measuring different aspects of the activation process, from upstream stimulus to downstream functional output.
Assay 1: Immunoblot Analysis of ATF6 Cleavage
This assay directly measures the proteolytic processing of full-length ATF6 (p90ATF6α, ~90 kDa) to its active, cytosolic fragment (p50ATF6α, ~50 kDa).
Detailed Protocol:
- Cell Treatment & Lysis: Treat cells (e.g., HEK293, HeLa) with ER stress inducers (e.g., 5 µg/mL Tunicamycin, 10 µM Thapsigargin) or test compounds for a time course (e.g., 1-8 hours). Use a modified RIPA buffer (with protease inhibitors) for whole-cell lysis. For nuclear enrichment, use a hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, protease inhibitors) followed by nuclear extraction with a high-salt buffer (20 mM HEPES, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA, 25% v/v glycerol).
- Gel Electrophoresis: Load 20-40 µg of protein per lane on a 4-12% Bis-Tris polyacrylamide gel. Include a pre-stained protein ladder.
- Membrane Transfer & Blocking: Transfer to a PVDF membrane using standard wet transfer. Block with 5% non-fat dry milk in TBST for 1 hour.
- Antibody Probing: Incubate with primary antibodies overnight at 4°C:
- Anti-ATF6α (e.g., ab122897): Detects both full-length and cleaved fragment.
- Anti-Lamin B1 (nuclear marker) and Anti-β-Tubulin (cytosolic marker) for fractionation validation. Wash and incubate with appropriate HRP-conjugated secondary antibodies.
- Detection: Develop using enhanced chemiluminescence (ECL) substrate and image.
Assay 2: Quantitative RT-PCR (qRT-PCR) of ATF6 Target Genes
Measures the transcriptional output of ATF6f, providing a functional readout of its activation.
Detailed Protocol:
- RNA Extraction: After treatment, lyse cells in TRIzol reagent and extract total RNA following the manufacturer's protocol. Treat with DNase I to remove genomic DNA contamination.
- cDNA Synthesis: Use 1 µg of total RNA with a reverse transcription kit (e.g., High-Capacity cDNA Reverse Transcription Kit) using random hexamers.
- qPCR Amplification: Prepare reactions with SYBR Green or TaqMan master mix. Primers/probes for canonical ATF6 targets include:
- HSPA5 (GRP78/BiP): Forward 5'-CCTGCGTCGGTGTGTTCAC-3', Reverse 5'-CAGTCGCTGGTACAGTTCCG-3'
- DERLIN3: Forward 5'-TGCTGGCTGTCATTGCTGTT-3', Reverse 5'-TCAGGAAGCTGAGGCAGAAG-3'
- XBP1 (spliced by IRE1, serves as UPR control): Forward 5'-CTGGAACAGCAAGTGGTAGA-3', Reverse 5'-CTGGATCAGACTGGGTGCTG-3' Normalize to housekeeping genes (e.g., GAPDH, ACTB). Run in triplicate on a real-time PCR system.
- Data Analysis: Calculate relative gene expression using the 2^(-ΔΔCt) method.
Assay 3: Immunofluorescence & Subcellular Localization
Visualizes the translocation of ATF6 from the ER to the Golgi and nucleus, a key step in its activation cascade.
Detailed Protocol:
- Cell Seeding and Treatment: Seed cells on poly-L-lysine-coated glass coverslips in a 24-well plate. Treat as required.
- Fixation and Permeabilization: Fix with 4% paraformaldehyde in PBS for 15 min at room temperature. Permeabilize with 0.2% Triton X-100 in PBS for 10 min.
- Blocking and Staining: Block with 5% BSA/1% goat serum in PBS for 1 hour. Incubate with primary antibodies (e.g., anti-ATF6α and anti-GM130 [Golgi marker] or anti-Calnexin [ER marker]) diluted in blocking buffer overnight at 4°C.
- Secondary Antibody & Mounting: Wash and incubate with fluorescent dye-conjugated secondary antibodies (e.g., Alexa Fluor 488, 594) and DAPI (for nuclei) for 1 hour in the dark.
- Imaging: Mount coverslips and image using a confocal or high-resolution fluorescence microscope. Co-localization analysis can be performed using software (e.g., ImageJ with Coloc2 plugin).
Assay 4: Luciferase Reporter Assay (ERSE-Luc)
Quantifies the transcriptional activity of ATF6f on a synthetic promoter containing ER stress response elements (ERSE).
Detailed Protocol:
- Reporter Transfection: Co-transfect cells in a 96-well plate with two plasmids:
- Reporter Plasmid: Firefly luciferase gene under the control of a promoter containing multiple ERSE sequences (e.g., p5xATF6-GL3).
- Control Plasmid: Renilla luciferase gene under a constitutive promoter (e.g., pRL-TK) for normalization.
- Treatment: 24-48 hours post-transfection, treat cells with ER stress inducers/test compounds.
- Luciferase Measurement: Lyse cells and measure Firefly and Renilla luciferase activities sequentially using a dual-luciferase reporter assay system on a microplate luminometer.
- Data Analysis: Calculate the ratio of Firefly/Renilla luminescence. Activity is expressed as fold-change over untreated control.
The following table summarizes expected outcomes from the described orthogonal assays in a canonical ER stress experiment using Tunicamycin (Tm) in HEK293 cells.
Table 1: Benchmarking ATF6 Activation Readouts Under ER Stress (Representative Data)
| Assay | Target/Metric | Basal (Vehicle) | Tunicamycin (5 µg/mL, 6h) | Key Interpretation |
|---|---|---|---|---|
| Immunoblot | p90ATF6α (FL) : p50ATF6α (frag) Ratio | High (e.g., 10:1) | Low (e.g., 1:2) | Cleavage and nuclear accumulation of ATF6f. |
| qRT-PCR | HSPA5 (GRP78) mRNA (Fold Change) | 1.0 ± 0.2 | 8.5 ± 1.5 | Functional transcriptional output of ATF6. |
| DERLIN3 mRNA (Fold Change) | 1.0 ± 0.3 | 12.0 ± 2.0 | Specific ATF6 target gene upregulation. | |
| Immunofluorescence | ATF6 Nuclear Localization (%) | <5% of cells | >70% of cells | Visual confirmation of activation cascade. |
| Luciferase Reporter | ERSE-Luc Activity (Fold Induction) | 1.0 ± 0.3 | 15.0 ± 3.0 | Direct measurement of ATF6f transcriptional activity. |
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagent Solutions for ATF6 Studies
| Reagent/Category | Example Product/Identifier | Primary Function in ATF6 Research |
|---|---|---|
| ER Stress Inducers | Tunicamycin, Thapsigargin, DTT, Brefeldin A | Induce ER stress, triggering the UPR and ATF6 activation pathway. |
| ATF6 Inhibitors | Ceapins, 4µ8C (IRE1 inhibitor, for specificity) | Pharmacologically inhibit ATF6 signaling; used for validation and control experiments. |
| Primary Antibodies | Anti-ATF6α (ab122897), Anti-GRP78/BiP (ab21685), Anti-Lamin B1, Anti-GM130 | Detect ATF6 protein forms, downstream targets, and organelle markers for localization. |
| Reporter Plasmids | p5xATF6-GL3 (Firefly), pRL-TK (Renilla) | Quantify ATF6 transcriptional activity via luminescence in reporter assays. |
| qPCR Assays | TaqMan Gene Expression Assays for HSPA5, DERLIN3, XBP1s | Pre-validated primers/probes for precise quantification of ATF6 target gene mRNA. |
| Cell Lines | HEK293, HeLa, MEFs (WT vs. Atf6α/β KO) | Standard models for UPR studies; KO lines provide essential negative controls. |
Signaling Pathway and Workflow Visualizations
ATF6 Activation Pathway Under ER Stress
Orthogonal Validation Workflow for ATF6
This analysis is situated within a broader thesis exploring the ATF6-GRP78 chaperone axis as a central regulatory node in proteostasis. While ATF6’s coupling to the master chaperone GRP78 is a focal point, a complete understanding of its role in protein folding requires a comparative examination of the two other UPR sensors: IRE1α and PERK. This guide provides a technical dissection of their activation mechanisms, signaling outputs, and functional cross-talk, providing a framework for targeted therapeutic intervention in protein misfolding diseases.
Quantitative Comparison of the Three UPR Arms
Table 1: Core Characteristics of ATF6, IRE1α, and PERK
| Feature | ATF6 (Activating Transcription Factor 6) | IRE1α (Inositol-Requiring Enzyme 1α) | PERK (PKR-like ER Kinase) |
|---|---|---|---|
| ER Luminal Sensor | GRP78 binding | GRP78 binding | GRP78 binding |
| Primary Activation Trigger | Dissociation of GRP78; Golgi trafficking | Dissociation of GRP78; dimerization/trans-autophosphorylation | Dissociation of GRP78; dimerization/trans-autophosphorylation |
| Key Effector Action | Site-1/2 protease cleavage; transcription factor release (ATF6f) | Unconventional splicing of XBP1 mRNA; RIDD | Phosphorylation of eIF2α |
| Major Transcriptional Target Genes | GRP78, GRP94, PDI, XBP1 | XBP1s targets (ERAD, lipid synthesis) | ATF4 targets (CHOP, amino acid metabolism, anti-oxidant response) |
| Primary Functional Focus | Chaperone upregulation & ER expansion | ERAD enhancement & ER expansion | Global translation attenuation; oxidative stress response |
| Approx. Activation Kinetics | Intermediate (hours) | Fast (minutes to hours) | Very Fast (minutes) |
| Phenotype of KO in Mice | Embryonic lethal (severe) | Embryonic lethal (severe) | Perinatal lethal (pancreatic dysfunction) |
Table 2: Outputs and Physiological Roles
| Pathway Output | ATF6 | IRE1α | PERK | Functional Overlap |
|---|---|---|---|---|
| ER Chaperone Induction | High (Direct) | Moderate (via XBP1s) | Low (via ATF4) | Cooperative ER folding capacity increase |
| ERAD Component Induction | Low | High (via XBP1s) | Moderate (via ATF4) | Synergistic clearance of misfolded proteins |
| Translation Attenuation | No | No | High (eIF2α-P) | PERK reduces influx; ATF6/IRE1 increase capacity |
| Apoptosis Promotion | Low (context-dependent) | Low (RIDD can promote) | High (sustained CHOP) | PERK/CHOP is major apoptotic arm |
| Lipid Synthesis | Yes | High (via XBP1s) | Indirect | IRE1-XBP1 central for ER membrane expansion |
Detailed Experimental Protocols for Key Assays
3.1. Monitoring ATF6 Activation & Processing (Immunoblot)
- Objective: To assess ATF6 activation via cleavage and nuclear translocation.
- Protocol:
- Cell Treatment & Lysis: Treat cells (e.g., HEK293) with ER stressor (e.g., 2µg/mL Tunicamycin, 5h). Prepare cytosolic and nuclear fractions using a commercial kit (e.g., NE-PER).
- Immunoblotting: Run 30-50µg protein/lane on 10% SDS-PAGE. Transfer to PVDF membrane.
- Detection: Probe with anti-ATF6 antibody (Clone 70B1413.1). The full-length protein (~90kDa) is ER-resident. The cleaved active fragment (ATF6f, ~50kDa) should be enriched in the nuclear fraction.
- Controls: Include β-actin (cytosolic) and Lamin B1 (nuclear) for fractionation purity. Use DTT (5mM, 1h) as a positive control for ATF6 activation.
3.2. Assessing IRE1α RNase Activity (XBP1 Splicing Assay)
- Objective: To detect IRE1α activation via its signature output, XBP1 mRNA splicing.
- Protocol:
- RNA Extraction: Extract total RNA from treated cells using TRIzol.
- RT-PCR: Perform reverse transcription. Amplify XBP1 cDNA using standard PCR with primers flanking the IRE1α cleavage site.
- Product Analysis: Run PCR products on a 2.5-3% agarose gel. Unspliced XBP1u yields a 473bp band; spliced XBP1s yields a 447bp band. For higher precision, use PstI restriction digest (XBP1u is cut, XBP1s is not).
- Quantification: Use qPCR with primers specific for XBP1s versus total XBP1.
3.3. Measuring PERK Activity (eIF2α Phosphorylation)
- Objective: To quantify PERK activation via its direct substrate.
- Protocol:
- Cell Lysis: Lyse cells in RIPA buffer containing phosphatase and protease inhibitors.
- Immunoblotting: Run proteins on 12% SDS-PAGE. Transfer to membrane.
- Dual Probing: Probe simultaneously or sequentially with:
- Anti-phospho-eIF2α (Ser51) antibody.
- Anti-total eIF2α antibody.
- Analysis: Calculate the p-eIF2α / total eIF2α ratio. Thapsigargin (1µM, 30min-1h) is an effective positive control.
Visualizations of UPR Signaling Pathways
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for UPR Pathway Analysis
| Reagent / Material | Primary Function in UPR Research | Example/Note |
|---|---|---|
| Pharmacological ER Stressors | Induce UPR activation controllably for experimentation. | Tunicamycin (N-glycosylation inhibitor). Thapsigargin (SERCA pump inhibitor). DTT (reducing agent causing ER protein misfolding). |
| Pathway-Specific Inhibitors | To dissect contributions of individual UPR arms. | GSK2606414 (potent PERK inhibitor). 4μ8c (IRE1α RNase domain inhibitor). Ceapins (selective ATF6 inhibitors). |
| Antibodies (Immunoblot/IF) | Detect protein expression, cleavage, phosphorylation, and localization. | Anti-ATF6 (full-length vs. cleaved). Anti-XBP1s (specific for spliced form). Anti-p-eIF2α (Ser51). Anti-GRP78/BiP. |
| Reporter Cell Lines | Real-time, quantitative readout of pathway activity. | ERSE or UPRE Luciferase reporters (ATF6/XBP1s activity). CHOP-luciferase (PERK/ATF4 output). FRET-based sensors for IRE1 activity. |
| qPCR Primer Panels | Quantify transcriptional output of all UPR arms. | Primer sets for: GRP78 (ATF6), EDEM1 (IRE1-XBP1s), CHOP (PERK-ATF4), and housekeeping genes (ACTB, GAPDH). |
| siRNA/shRNA Libraries | For targeted knockdown of UPR components to study loss-of-function. | Validated pools targeting ATF6, ERN1 (IRE1), EIF2AK3 (PERK), XBP1. |
| CRISPR/Cas9 KO Cell Lines | Generate stable, complete knockout models for mechanistic studies. | Isogenic cell lines lacking ATF6, IRE1α, or PERK to study compartment-specific functions and redundancy. |
The unfolded protein response (UPR) is a critical cellular adaptive mechanism activated by endoplasmic reticulum (ER) stress. The ATF6 (Activating Transcription Factor 6) branch is a key sensor and transducer, which, under stress, traffics to the Golgi to be cleaved. Its cytosolic fragment then translocates to the nucleus to upregulate chaperone genes, including the master ER chaperone GRP78/BiP. Validating the precise in vivo functions, compensatory mechanisms, and therapeutic potential of this axis necessitates robust genetic interrogation. This guide details the core animal model strategies—knockout (KO), knockdown (KD), and transgenic (TG)—for the genetic validation of components within the ATF6-GRP78 system, providing a technical framework for protein folding researchers.
Core Genetic Validation Strategies: Methodologies and Applications
Knockout Models: Constitutive and Conditional
Objective: To completely and permanently abolish gene function, revealing non-redundant roles and systemic phenotypes.
- Protocol for Conventional Global KO (e.g., Atf6α -/-): The standard method employs homologous recombination in embryonic stem (ES) cells.
- Targeting Vector Construction: Design a vector with sequences homologous to the Atf6 gene flanks, replacing a critical exon (e.g., containing the DNA-binding domain) with a positive selection marker (e.g., neomycin resistance gene, Neo^r).
- ES Cell Electroporation & Selection: Introduce the vector into mouse ES cells via electroporation. Select successfully transfected cells with G418 (neomycin analog).
- Screening & Blastocyst Injection: Screen ES cell clones via PCR and Southern blot for correct homologous recombination. Inject validated ES cells into mouse blastocysts to generate chimeric founders.
- Germline Transmission & Breeding: Breed chimeras to wild-type mice to achieve germline transmission. Intercross heterozygous offspring to generate homozygous global KO mice.
- Protocol for Conditional KO (e.g., Grp78 floxed): Utilizes Cre-loxP technology for spatiotemporal control.
- "Floxed" Allele Creation: Generate a mouse line where exons critical for Grp78 function are flanked by loxP sites (floxed) via homologous recombination, without disrupting normal gene function.
- Crossing with Cre Drivers: Cross the floxed mouse with a line expressing Cre recombinase under a tissue-specific (e.g., Alb-Cre for hepatocytes) or inducible (e.g., Cre-ERT2 with tamoxifen) promoter.
- Gene Deletion: Cre expression catalyzes recombination between loxP sites, excising the floxed exons and creating a null allele in the target cells/tissue at the desired time.
Knockdown Models: Transient and Stable
Objective: To achieve partial, reversible gene silencing, mimicking hypomorphic alleles or allowing assessment of acute inhibition.
- Protocol for In Vivo siRNA/shRNA Delivery:
- Design & Validation: Design siRNA sequences targeting Atf6 mRNA or shRNA constructs for viral delivery. Validate silencing efficiency (>70%) and specificity in vitro (e.g., in hepatocyte cell lines) via qRT-PCR and immunoblotting.
- Formulation & Administration: For systemic delivery, formulate validated siRNAs with lipid nanoparticles (LNPs). For local delivery (e.g., brain), use stereotactic injection of lentiviral or AAV vectors encoding the shRNA.
- Dosing & Timing: Administer a single dose (e.g., 3 mg/kg siRNA IV) or multiple doses. Analyze tissues for mRNA/protein knockdown and phenotypic consequences (e.g., sensitivity to tunicamycin-induced ER stress) 3-7 days post-treatment.
Transgenic Models: Overexpression and Reporter
Objective: To study gain-of-function, rescue phenotypes, or visualize gene expression patterns in real-time.
- Protocol for ATF6 Constitutively Active (ATF6-N) Transgenic:
- Transgene Construction: Clone the cDNA encoding the cleaved, nuclear-active fragment of ATF6 (ATF6-N) downstream of a strong, ubiquitous promoter (e.g., CAG) or a tissue-specific promoter (e.g., Pdx1 for pancreatic β-cells).
- Pronuclear Microinjection: Purify the linearized transgene construct and microinject it into the pronucleus of fertilized mouse oocytes.
- Founder Identification & Line Establishment: Implant oocytes into pseudo-pregnant females. Screen offspring (founders) for transgene integration by genomic PCR and expression by immunoblot. Establish stable transgenic lines from founders with high expression.
- Protocol for GRP78 Reporter Mouse (e.g., Grp78-GFP):
- Knock-in Strategy: Using homologous recombination in ES cells, insert a GFP (or luciferase) reporter cassette, preceded by an internal ribosome entry site (IRES) or a self-cleaving 2A peptide, into the 3' UTR of the endogenous Grp78 gene, just before the stop codon.
- Validation: This ensures GFP expression is directly coupled to endogenous Grp78 transcription. Validate that the knock-in allele does not disrupt normal GRP78 protein function.
Comparative Data from ATF6/GRP78 Studies
Table 1: Phenotypic Outcomes from Genetic Manipulation of the ATF6-GRP78 Axis in Mice
| Genetic Model | Target Gene | Key Phenotypic Outcome | Quantitative Data (Example) |
|---|---|---|---|
| Constitutive Global KO | Atf6α | Viable, but severe embryonic lethality under in utero ER stress. Impaired glucose homeostasis. | Embryonic survival after tunicamycin (TM): WT: 85%, KO: <10% (P<0.001). |
| Conditional KO (Liver) | Grp78 | Spontaneous hepatosteatosis, increased sensitivity to high-fat diet induced ER stress and injury. | Serum ALT levels (HFD, 12wks): Control: 35 U/L, KO: 120 U/L (P<0.01). |
| Transgenic Overexpression | ATF6-N (Pancreas) | Protection against β-cell apoptosis in diabetic models, improved insulin secretion. | β-cell apoptosis in db/db mice: Non-TG: 4.5%, TG: 1.2% (P<0.05). |
| AAV-mediated KD (Brain) | Atf6 | Exacerbated neuronal loss and behavioral deficits in a model of neurodegenerative disease (e.g., Parkinson's). | Dopaminergic neurons surviving: Scramble-shRNA: 65%, Atf6-shRNA: 38% (P<0.005). |
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Reagents for Genetic Validation of the ATF6-GRP78 System
| Reagent / Material | Function / Application |
|---|---|
| CRISPR-Cas9 Ribonucleoprotein (RNP) | For rapid generation of KO/KI models via direct microinjection into zygotes, bypassing ES cells. |
| Tamoxifen | Inducer for Cre-ERT2 systems; enables temporal control of conditional KO or reporter activation in adult animals. |
| Tunicamycin | N-linked glycosylation inhibitor; standard pharmacological agent to induce acute ER stress in vivo for challenge tests. |
| Adeno-Associated Virus (AAV) Serotype 9 | Efficient vector for in vivo shRNA delivery or transgene overexpression with broad tissue tropism (liver, heart, CNS). |
| Lipid Nanoparticles (LNPs) | Formulation for systemic, in vivo delivery of siRNA oligonucleotides targeting Atf6 or Grp78. |
| Anti-GRP78/BiP Antibody (Clone C50B12) | Validated antibody for immunoblotting and immunohistochemistry to monitor GRP78 protein levels in tissues. |
| ER-Tracker Dyes | Live-cell imaging dyes to assess ER morphology and stress in primary cells isolated from genetically modified models. |
Visualizing Experimental Workflows and Pathways
Diagram Title: Genetic Model Validation Workflow
Diagram Title: ATF6-GRP78 ER Stress Signaling Pathway
Within the broader thesis on the ATF6/GRP78 chaperone system in protein folding research, the Unfolded Protein Response (UPR) represents a critical cellular homeostasis network. Among its three arms—PERK, IRE1α, and ATF6—the ATF6 pathway, with its central chaperone GRP78/BiP, offers unique therapeutic leverage points for diseases characterized by proteostasis imbalance, including neurodegenerative disorders, cancer, and metabolic diseases. This whitepaper provides a technical comparison of the therapeutic targeting landscape, focusing on small molecules and biologics for the ATF6/GRP78 axis versus modulators of the PERK and IRE1α pathways.
Title: UPR Signaling Pathways Initiated by GRP78 Dissociation
Comparative Therapeutic Targeting Landscape
Table 1: Small Molecule Modulators of UPR Pathways
| Target/Pathway | Compound Name (Example) | Mechanism of Action | Development Stage (as of 2024) | Key Disease Indications |
|---|---|---|---|---|
| ATF6 Activation | AA147 [cite:PMID 36368609] | Activates ATF6 via selective covalent modification of ER-resident dicarbonyl/l-xylulose reductase (DCXR). | Preclinical | Neurodegeneration, Ischemic Injury |
| ATF6 Activation | Ceapins (e.g., Ceapin-A7) [cite:PMID 29925948] | Selective ATF6 activators by inhibiting its transport from ER to Golgi. | Preclinical (Tool Compound) | Proteostasis Diseases |
| GRP78 Inhibition | HA15 | Binds GRP78's ATPase domain, inhibits activity, induces ER stress. | Preclinical | Cancer (Melanoma, Glioblastoma) |
| GRP78 Cell Surface | Patrimoine 3 | Monoclonal antibody against cell-surface GRP78. | Preclinical | Cancer |
| PERK Inhibition | GSK2606414, AMG PERK 44 | ATP-competitive PERK kinase inhibitors. | Phase I (Discontinued) | Neurodegeneration |
| PERK Modulation | ISRIB | Reverses effects of p-eIF2α, downstream of PERK. | Preclinical/Research | Cognitive Deficits |
| IRE1α Inhibition | 4μ8C, MKC-3946 | Inhibits IRE1α RNase activity. | Preclinical (4μ8C), Phase I (MKC-3946, halted) | Multiple Myeloma, Cancer |
| IRE1α Modulation | STF-083010 | Selective IRE1α endoribonuclease inhibitor. | Preclinical | Cancer |
Table 2: Biologics and Other Modalities Targeting UPR
| Modality | Target | Agent Name/Type | Mechanism/Therapeutic Goal | Stage |
|---|---|---|---|---|
| Monoclonal Antibody | Cell-surface GRP78 | MAb159 [cite:PMID 31019024] | Blocks oncogenic signaling, induces internalization. | Preclinical/Phase I |
| Monoclonal Antibody | IRE1α | Anti-IRE1α (e.g., 64B12 mAb) | Blocks XBP1 splicing, induces IRE1α degradation. | Preclinical |
| Proteolysis Targeting Chimera (PROTAC) | PERK | PERK PROTACs (e.g., LC-2) | Induces selective ubiquitination and degradation of PERK. | Preclinical (Research) |
| Gene Therapy | ATF6 | AAV-encoded active ATF6 | Direct delivery of transcriptionally active ATF6 fragment. | Preclinical (in vivo proof-of-concept) |
| Antisense Oligo (ASO) | XBP1s | XBP1s ASO | Reduces levels of spliced XBP1 mRNA. | Preclinical |
Table 3: Quantitative Comparison of Key UPR Modulator Effects (Representative Preclinical Data)
| Compound | Target | Key Readout (Cell-Based Assay) | Reported Efficacy (IC50/EC50) | Model System |
|---|---|---|---|---|
| AA147 | ATF6 Activator | ATF6-luciferase reporter, GRP78 mRNA | EC50 ~3.5 μM | HEK293T, Primary Neurons |
| Ceapin-A7 | ATF6 Activator | Nuclear ATF6(p50) localization | Active at 10 μM | HeLa, MEFs |
| HA15 | GRP78 Inhibitor | Cell Viability (MTT), Caspase-3/7 activity | IC50 ~1-4 μM (viability, melanoma) | A375 Melanoma, Patient-derived cells |
| GSK2606414 | PERK Inhibitor | PERK autophosphorylation (pPERK) | IC50 = 0.4 nM | PC12, Mouse Brain Homogenate |
| 4μ8C | IRE1α RNase Inhibitor | XBP1 splicing (RT-PCR) | IC50 = 8.9 μM | Multiple Myeloma Cells |
Key Experimental Protocols
Protocol: ATF6 Luciferase Reporter Assay for Compound Screening
Purpose: Quantify ATF6 pathway activation by candidate small molecules. Reagents: HEK293T cells, pGL4-ERSE (ER Stress Response Element)-luciferase plasmid, pRL-TK Renilla plasmid, Lipofectamine 3000, Dual-Luciferase Reporter Assay System, test compounds (e.g., AA147, thapsigargin as positive control). Procedure:
- Day 1: Seed HEK293T cells in 96-well white plates at 20,000 cells/well.
- Day 2: Co-transfect each well with 100 ng pGL4-ERSE-luciferase and 10 ng pRL-TK Renilla using Lipofectamine 3000 per manufacturer's protocol.
- Day 3: Treat cells with serially diluted compounds or vehicle (DMSO, <0.1%) for 12-16 hours.
- Day 4: Lyse cells and measure firefly and Renilla luciferase activity sequentially using the Dual-Luciferase Assay on a plate reader. Normalize firefly luminescence to Renilla luminescence for each well. Plot normalized luciferase activity vs. compound concentration to determine EC50.
Protocol: Monitoring GRP78 Release via Co-Immunoprecipitation
Purpose: Validate direct GRP78 inhibition or dissociation from UPR sensors. Reagents: Cell line of interest (e.g., A375), RIPA lysis buffer, protease/phosphatase inhibitors, anti-GRP78 antibody, protein A/G agarose beads, SDS-PAGE gel, antibodies for immunoblotting (anti-ATF6, anti-IRE1α, anti-PERK). Procedure:
- Treat cells with compound (e.g., HA15) or ER stressor (Tunicamycin 2 μg/mL, 6h).
- Lyse cells in ice-cold RIPA buffer with inhibitors. Centrifuge at 14,000g, 15 min, 4°C.
- Pre-clear lysate with protein A/G beads for 30 min at 4°C.
- Incubate 500 μg of pre-cleared lysate with 2 μg of anti-GRP78 antibody overnight at 4°C.
- Add protein A/G beads for 2h. Pellet beads and wash 3x with lysis buffer.
- Elute bound proteins in 2X Laemmli buffer at 95°C for 5 min.
- Perform SDS-PAGE and immunoblot for ATF6, IRE1α, and PERK. Increased co-precipitation of sensors with GRP78 indicates inhibition of stress-induced dissociation.
Protocol: XBP1 Splicing Assay (IRE1α Activity)
Purpose: Assess IRE1α RNase inhibition by compounds like 4μ8C. Reagents: TRIzol, High-Capacity cDNA Reverse Transcription Kit, PCR primers flanking the IRE1α splice site (human: F: 5'-CCTGGTTGCTGAAGAGGAGG-3', R: 5'-CCATGGGAAGATGTTCTGGG-3'), standard PCR mix, agarose gel. Procedure:
- Treat cells with IRE1α modulator (e.g., 4μ8C at 50 μM) ± tunicamycin (2 μg/mL, 4h).
- Extract total RNA with TRIzol and synthesize cDNA.
- Perform PCR (30-35 cycles) using primers above. PCR product for unspliced XBP1 (XBP1u) = 289 bp, spliced XBP1 (XBP1s) = 263 bp.
- Resolve PCR products on a 2.5-3% agarose gel. Inhibition of IRE1α reduces the intensity of the lower (spliced) band under stress conditions.
Title: Workflow for Identifying and Validating ATF6/GRP78-Targeted Compounds
The Scientist's Toolkit: Key Research Reagent Solutions
Table 4: Essential Reagents for ATF6/GRP78 and UPR Research
| Reagent Category | Specific Item/Assay | Function & Application | Example Vendor(s) |
|---|---|---|---|
| Cell-Based Reporters | ATF6 Reporter Kit (Luciferase) | Quantifies ATF6 pathway activation in live cells for HTS. | Promega, Indicia Biotechnology |
| Cell-Based Reporters | XBP1 Splicing Reporter (GFP/RFP) | Visualizes IRE1α activity via fluorescence shift upon splicing. | Addgene (plasmid #64936) |
| Key Antibodies | Anti-GRP78/BiP (Monoclonal) | Immunoblot, IP, IHC to monitor GRP78 expression and localization. | Cell Signaling Tech (3177S), Abcam (ab21685) |
| Key Antibodies | Anti-ATF6 (p50) (Polyclonal) | Detects cleaved, active ATF6 fragment in nucleus via immunoblot/IF. | Novus Biologicals (NBP1-40256) |
| Key Antibodies | Anti-p-PERK (Thr980) | Monitors PERK activation status via immunoblot. | Cell Signaling Tech (3179S) |
| Chemical Inducers/Inhibitors | Tunicamycin, Thapsigargin | Standard ER stress inducers (positive controls). | Sigma-Aldrich, Tocris |
| Chemical Inducers/Inhibitors | AA147, Ceapin-A7 | Tool compounds for selective ATF6 pathway activation. | Custom synthesis, Sigma (Ceapin-A7: SML2463) |
| Chemical Inducers/Inhibitors | GSK2606414, 4μ8C | Tool compounds for PERK and IRE1α inhibition, respectively. | Tocris, Sigma-Aldrich |
| Detection Kits | Dual-Luciferase Reporter Assay System | Sensitive quantification of reporter gene activity. | Promega |
| Detection Kits | RT-qPCR Assays for UPR Targets | Quantifies mRNA levels of GRP78, CHOP, XBP1s, etc. | Qiagen, Thermo Fisher (TaqMan) |
| Critical Cell Lines | WT and ATF6-/-, IRE1α-/- MEFs | Isogenic cell pairs for validating target specificity of compounds. | ATCC, academic repositories |
The ATF6/GRP78 axis presents a distinct and promising therapeutic node within the UPR, with emerging small-molecule activators like AA147 offering a novel strategy to pre-emptively boost proteostasis capacity. In contrast, PERK and IRE1α inhibitors largely aim to mitigate maladaptive UPR signaling in chronic disease. The choice of target depends fundamentally on the disease context—whether the therapeutic goal is to enhance adaptive UPR (via ATF6) or suppress terminal UPR (via PERK/IRE1α). Continued development of selective tool compounds and biologics, coupled with rigorous mechanistic protocols as outlined, is essential to delineate their full translational potential within protein folding research and beyond.
This whitepaper situates the assessment of Activating Transcription Factor 6 (ATF6) pathway activity within the broader thesis on the ATF6-GRP78 chaperone system in protein folding research. The unfolded protein response (UPR) sensor ATF6 and its central downstream effector, the chaperone GRP78/BiP, constitute a critical adaptive mechanism for endoplasmic reticulum (ER) proteostasis. Dysregulation of this axis is implicated in a spectrum of diseases, from neurodegeneration to cancer. Consequently, quantifiable measures of ATF6 activation in human biospecimens transition from being fundamental research tools to possessing significant diagnostic and prognostic biomarker potential. This guide provides a technical framework for such assessment.
Core Signaling Pathway: ATF6 Activation and Regulation
The ATF6 pathway is a tightly regulated cascade initiated by ER stress.
Diagram Title: ATF6 Activation Pathway from ER Stress to Gene Transcription
Key Biomarkers of ATF6 Pathway Activity
Quantitative data on ATF6 pathway components can be categorized as direct or indirect biomarkers.
Table 1: Core ATF6 Pathway Biomarkers in Human Samples
| Biomarker Category | Specific Target | Detection Method | Biological Significance | Correlation with Pathway Activity |
|---|---|---|---|---|
| Direct | ATF6 p90 (Full-length) | WB, ELISA | Inactive, ER-resident precursor | Negative |
| ATF6 p50 (Cleaved) | WB, IF (Nuclear) | Active transcription factor | Positive | |
| ATF6 Target Gene mRNA (GRP78, CHOP) | qRT-PCR, RNA-seq | Transcriptional output | Positive | |
| Indirect / Surrogate | GRP78/BiP Protein | IHC, ELISA, WB | Primary chaperone, negative regulator | Often positive (feedback) |
| sXBP1 mRNA Splicing | qRT-PCR (Assay) | IRE1α pathway activity; often co-activated | Context-dependent | |
| ERSE/UPRE Luciferase Reporter | Ex vivo assay (Cell line) | Integrated UPR element activity | Positive |
Table 2: Exemplary Prognostic Correlations in Human Cancers (Recent Findings)
| Cancer Type | Sample Type | Key ATF6/GRP78 Finding | Prognostic Value | Reported Hazard Ratio (Approx.) |
|---|---|---|---|---|
| Glioblastoma | Tumor Tissue (IHC) | High nuclear ATF6 (p50) & GRP78 | Poor Overall Survival | 2.1 (95% CI: 1.3-3.4) |
| Hepatocellular Carcinoma | Serum (ELISA) | Elevated soluble GRP78 | Tumor Stage Correlation | p<0.001 |
| Multiple Myeloma | Bone Marrow (qRT-PCR) | High GRP78 & ATF6 mRNA | Resistance to Proteasome Inhibitors | Progression-Free Survival: HR 1.8 |
| Breast Cancer | Tumor Tissue (WB/IHC) | ATF6 Cleavage Ratio (p50/p90) | Metastasis & Recurrence | p=0.007 |
Experimental Protocols for Human Sample Analysis
Protocol: Immunoblotting for ATF6 Cleavage Status in Tissue Lysates
Objective: Quantify the ratio of active (p50) to inactive (p90) ATF6.
Key Reagents:
- RIPA Lysis Buffer (with protease inhibitors)
- Anti-ATF6 Antibody (clone 70B1413.1 or polyclonal ab37149)
- HRP-conjugated secondary antibody
- Enhanced Chemiluminescence (ECL) substrate
- Pre-cast gradient (4-20%) polyacrylamide gels
Procedure:
- Sample Preparation: Homogenize 20-30 mg of frozen tissue in 300 µL ice-cold RIPA buffer. Centrifuge at 16,000 x g for 15 min at 4°C. Collect supernatant.
- Protein Quantification: Use BCA assay. Load 30-50 µg total protein per lane.
- Electrophoresis & Transfer: Run on SDS-PAGE gel. Transfer to PVDF membrane using standard wet transfer.
- Immunodetection: Block membrane with 5% non-fat milk in TBST for 1h. Incubate with primary anti-ATF6 antibody (1:1000) overnight at 4°C. Wash 3x with TBST. Incubate with HRP-secondary (1:5000) for 1h at RT. Develop using ECL and image with chemiluminescence detector.
- Analysis: Quantify band intensities for p90 (~90 kDa) and p50 (~50 kDa) using ImageJ. Calculate p50/p90 ratio.
Protocol: qRT-PCR for ATF6 Target Gene Expression from Blood RNA
Objective: Measure mRNA levels of canonical ATF6 targets (GRP78/HSPA5, CHOP/DDIT3).
Key Reagents:
- PAXgene Blood RNA tubes or TRIzol LS
- High-Capacity cDNA Reverse Transcription Kit
- TaqMan Gene Expression Assays (e.g., HSPA5: Hs00607129gH, *DDIT3*: Hs00358796g1, ACTB: Hs01060665_g1)
- Real-Time PCR System-compatible plates.
Procedure:
- RNA Isolation: Isolve total RNA from whole blood per manufacturer's protocol. Include DNase I treatment. Assess RNA integrity (RIN >7).
- cDNA Synthesis: Convert 1 µg total RNA to cDNA using random hexamers and MultiScribe Reverse Transcriptase.
- Quantitative PCR: Perform triplicate reactions with 10 ng cDNA equivalent, TaqMan Master Mix, and specific assay. Use standard cycling conditions (95°C for 10 min, followed by 40 cycles of 95°C for 15s and 60°C for 1 min).
- Data Analysis: Calculate ∆Ct values relative to housekeeping gene (e.g., ACTB). Use the comparative ∆∆Ct method for relative quantification across sample groups.
Protocol: Immunohistochemistry for GRP78 and Nuclear ATF6 in FFPE Sections
Objective: Spatially localize and semi-quantify protein expression in tumor microenvironment.
Key Reagents:
- FFPE tissue sections (4 µm)
- Antigen retrieval buffer (pH 6.0 citrate or pH 9.0 EDTA)
- Anti-GRP78 antibody (clone C50B12) and anti-ATF6 p50 antibody
- HRP-polymer detection system (e.g., EnVision+)
- DAB chromogen and hematoxylin counterstain.
Procedure:
- Deparaffinization & Retrieval: Bake slides, deparaffinize in xylene, rehydrate through graded alcohols. Perform heat-induced epitope retrieval in appropriate buffer for 20 min.
- Staining: Block endogenous peroxidases with 3% H₂O₂. Apply protein block (serum), then primary antibody (GRP78 1:200; ATF6 p50 1:100) for 1h at RT or overnight at 4°C.
- Detection & Visualization: Apply HRP-labeled polymer for 30 min. Develop with DAB for 5-10 min. Counterstain with hematoxylin.
- Scoring: Use semi-quantitative H-score (H-SCORE = Σ (pi × i), where pi is % of cells stained at intensity i (0-3)). Assess nuclear (ATF6 p50) and cytoplasmic/membrane (GRP78) staining separately.
Integrated Workflow for Biomarker Assessment
Diagram Title: Workflow for ATF6 Pathway Biomarker Assessment from Human Samples
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents for ATF6 Pathway Biomarker Research
| Reagent Category | Specific Item/Kit | Primary Function in ATF6 Research |
|---|---|---|
| Antibodies | Anti-ATF6 (Full length, p90) | Detects inactive ER-resident precursor via WB/IHC. |
| Anti-ATF6 (Cleaved, p50) | Specific detection of active nuclear transcription factor. | |
| Anti-GRP78/BiP (Multiple clones) | Gold-standard chaperone marker; IHC, WB, IP. | |
| Assay Kits | Human GRP78/BiP ELISA Kit | Quantifies soluble GRP78 in serum/plasma/culture supernatant. |
| TaqMan UPR Stress Panel | Multi-gene qPCR array for ATF6, IRE1, PERK targets. | |
| ER Stress Reporter (Luciferase) | Cell-based assay for UPRE/ERSE activity. | |
| Chemical Inducers/Inhibitors | Thapsigargin | SERCA inhibitor; robust inducer of ER stress and ATF6 activation. |
| Tunicamycin | N-glycosylation blocker; induces ER stress. | |
| AEBSF hydrochloride | Site-1 Protease (S1P) inhibitor; blocks ATF6 cleavage. | |
| Sample Prep | RIPA Lysis Buffer (with inhibitors) | Comprehensive protein extraction for WB analysis. |
| PAXgene Blood RNA System | Stabilizes blood RNA for transcriptomic analysis from whole blood. | |
| FFPE RNA/DNA Extraction Kit | Isolves nucleic acids from archived formalin-fixed tissue. |
Conclusion
The ATF6-GRP78 chaperone system represents a sophisticated and druggable nexus within the cellular proteostasis network. This article has detailed its foundational mechanisms, practical study methodologies, optimization strategies, and validation frameworks. Key takeaways include its unique role as a sensor and transcriptional master regulator of ER chaperones, the critical need for specific tools to disentangle its activity from parallel UPR branches, and its promising, yet complex, therapeutic potential. Future directions must focus on developing highly specific ATF6 modulators, understanding temporal and tissue-specific pathway dynamics, and translating mechanistic insights into clinical strategies for diseases of protein misfolding, such as Alzheimer's, diabetes, and certain cancers, where restoring ER balance could halt progression.